Data Science Explored a space to learn and analyze
Predicting the Presence of Heart Disease in Patients using Python
The usefulness of being able to accurately anticipate and predict the presence of heart disease cannot be understated. Heart disease is the world’s leading cause of death for both men and women 1.
- Approximately 647,000 American lives are lost each year to the disease - accounting for one in every four U.S. deaths 2,3.
- The cost of heart disease in the United States, from 2014 to 2015, totaled $219 billion 3. This included the cost of health care services such as tests and procedures, medicines, and lost productivity.
This project details the process I took to build a model that is capable of predicting the presence of heart disease in patients as well as outlining ways to use it in order to combat the devastation caused by the disease.
Please leave a comment at the bottom of the page to let me know your thoughts on the project and the actions I took to arrive at the final model. At the end of the Project Overview, there are questions posed for reflection and deliberation - feel free to answer one of those too if you would like.
Project Summary
- Statistical analysis, data mining techniques, and five machine learning models were built and ensembled to accurately predict the presence of heart disease in patients from the Hungarian Institute of Cardiology in Budapest.
- The model that provided the optimal combination of total patients predicted correctly and F1 Score, while being the most parsimonious, was the Support Vector Machine Classification Model #4. It was able to correctly predict the presence, or lack thereof, of heart disease in 86% of patients.
A summary of this models results can be seen directly below, and a full summary of all sixteen models can be found in the Model Visualization, Comparison, and Selection section.
| Model | F1 Score | Recall | Precision | Total Correct | Total Incorrect |
|---|---|---|---|---|---|
| Support Vector Machine Classifier Four | 0.804 | 0.774 | 0.837 | 252 | 40 |
Code snippets will be provided for each section outlined in the Project Overview at the bottom of the page. The snippets will encompass the entire script, broken into their related sections. If you would like to view the code script in its entirety, please visit this link.
Project Overview
i. Data Ingestion
ii. Data Cleaning
iii. Exploratory Data Analysis
iv. Model Building
v. Model Visualization, Comparison, and Selection
vi. Visualize Best Model
vii. Model Usefulness
viii.Questions for Consideration
Data Ingestion (View code)
The first step was obtaining the data and data dictionary from the UCI Machine Learning Repository 4. The files were saved in an appropriate location and then read into Python.
Data Cleaning (View code)
After the data was properly read into Python and the appropriate column names were supplied, data cleaning was performed. This involved:
- Removing unnecessary columns
- Converting column types
- Correcting any data discrepancies
- Removing patients with a large percentage of missing values
- In this particular case, large meant 10%. As a result, two patients were removed from the data set.
- Imputing missing values for patients using K-Nearest Neighbors, an advanced data imputation method
- Setting the target variable (“num”) to a binary range as previous studies have done
Exploratory Data Analysis (View code)
The following three images provide a sample of the analysis performed.
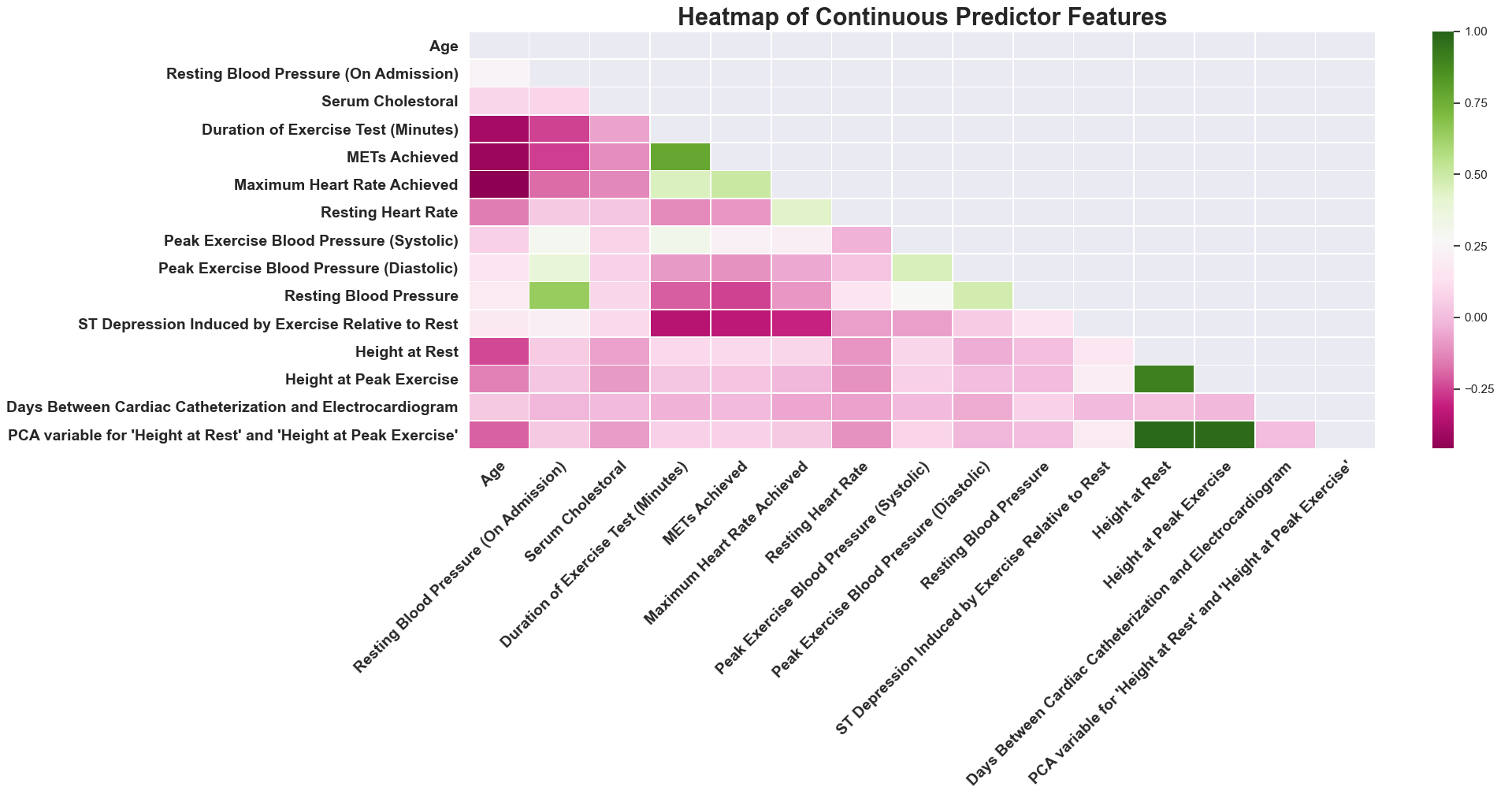
This heatmap shows correlation coefficients between each of the initial continuous variables plus two features, “Days Between Cardiac Catheterization and Electrocardiogram” and “PCA variable for ‘Height at Rest’ and ‘Height at Peak Exercise’”, created in the early stages of feature engineering. This visualization gives you information that is useful in performing data transformations and further feature engineering. Each unique cell, with its color, helps to provide information on the strength of the linear relationship between the two variables that interact to make it up.
- Take, as an example, the cell that is made up of ‘METs Achieved’ and ‘Duration of Exercise Test (Minutes).’ The darker green color indicates a positive correlation which implies a postive relationship between the two variables. So as ‘METs Achieved’ goes up, ‘Duration of Exercise Test (Minutes)’ has a tendency of going up.
- For an example of a negative correlation between two features, observe ‘Maximum Heart Rate Achieved’ and ‘Age.’ The dark pink color that makes up this cell suggests a negative correlation - this implies a negative relationship between ‘Maximum Heart Rate Achieved’ and ‘Age.’ As ‘Maximum Heart Rate Achieved’ goes up, ‘Age’ has a tendency of going down.
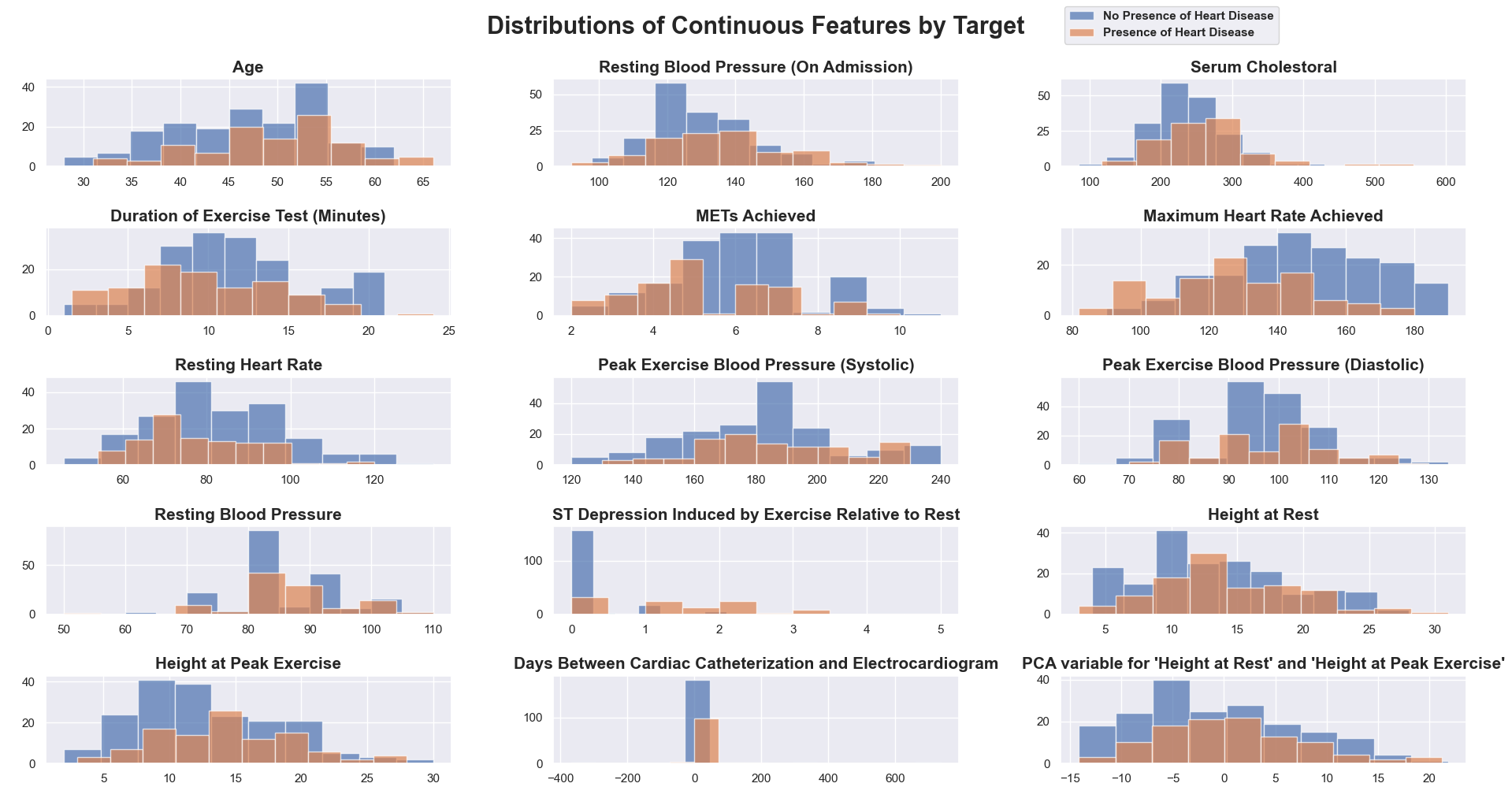
Above, there is a histogram for each of the initial continuous features against the target variable (diagnosis of heart disease) plus the two features created in the early stages of feature engineering, “Days Between Cardiac Catheterization and Electrocardiogram” and “PCA variable for ‘Height at Rest’ and ‘Height at Peak Exercise’.” This visualization allows you to see which of the predictor variables have noticeable differences in their distributions when split on the target, and would therefore be useful in prediction.
- A solid example of this is ‘Maximum Heart Rate Achieved.’ The blue histogram, which makes up the patients who have no presence of heart disease, is shifted to the right along the x-axis. The majority of those patients are able to achieve a higher maximum heart rate than the patients who do have a presence of heart disease (the orange histogram).
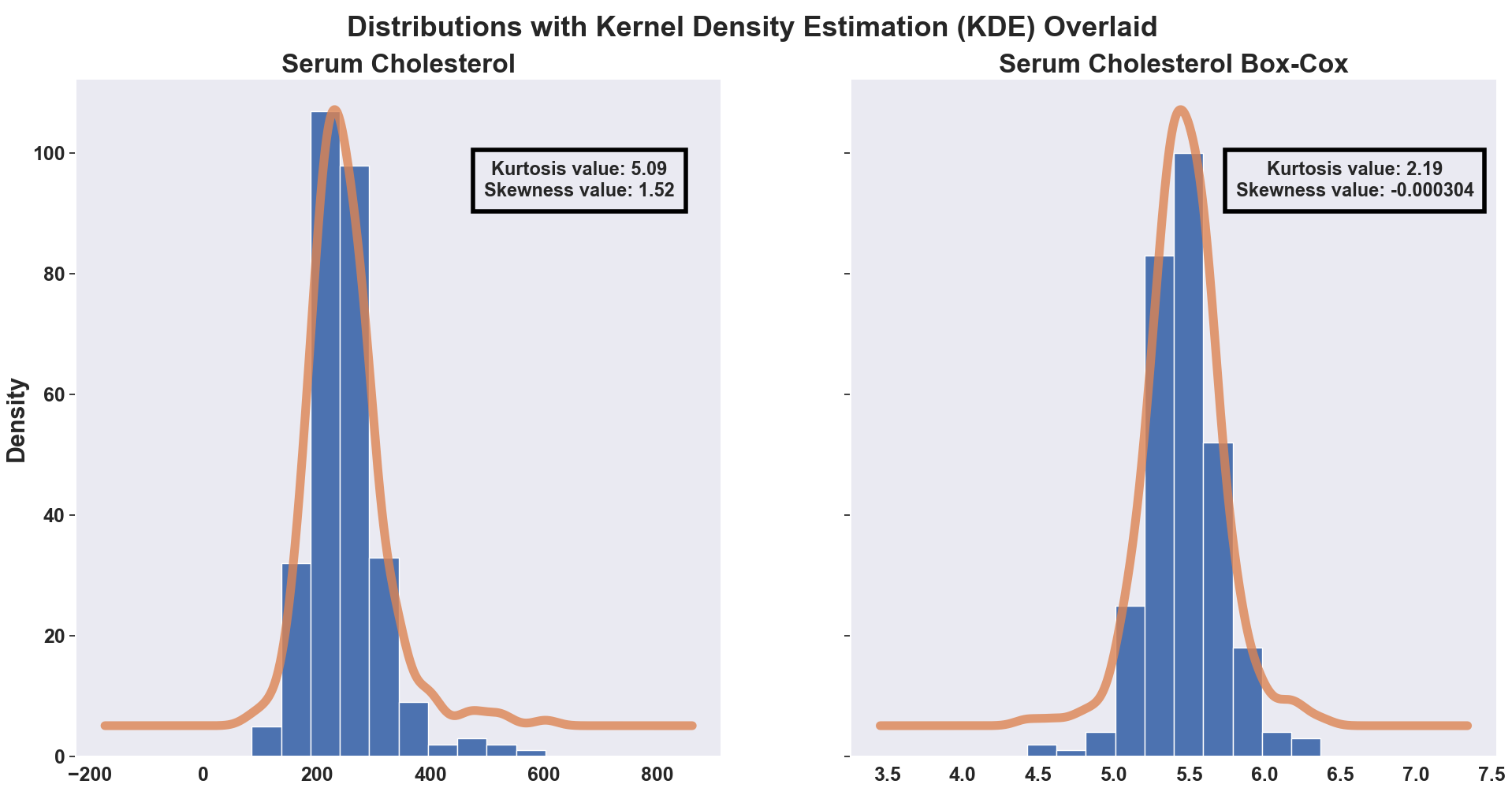
The above histograms show Serum Cholesterol on the left with no data transformation performed and on the right with a Box-Cox transformation applied.
- Regarding the left histogram:
- Notice the high, positive kurtosis value (please note this value is an adjusted version of Pearson’s kurtosis, known as excess kurtosis, where three is subtracted from the original kurtosis value to provide the comparison to a normal distribution - a value of 0.0 would be the excess kurtosis value of a normal distribution) labeled on the left histogram.
- This is a leptokurtotic distribution, meaning there is more data in these tails than in the tails of a normal distribution.
- The positive skewness value indicates a right-tailed distribution (a value of 0.0 would indicate a normal distribution).
- The kernel density estimation, which can be thought of as a locally smoothed version of the histogram, is overlaid to help visualize the shape. Both components, the lepotokurtotic distribution and right-tailed distribution, can be seen in the visualization with the help of the overlaid kernel density estimation.
- Notice the high, positive kurtosis value (please note this value is an adjusted version of Pearson’s kurtosis, known as excess kurtosis, where three is subtracted from the original kurtosis value to provide the comparison to a normal distribution - a value of 0.0 would be the excess kurtosis value of a normal distribution) labeled on the left histogram.
- The histogram to the right shows Serum Cholesterol with a Box-Cox transformation performed.
- Notice the much lower kurtosis value, although still positive and representing a leptokurtotic distribution.
- The skewness value is now nearly zero, representing much more normally distributed data.
Comparing the two histograms, it is evident the Box-Cox transformation was helpful in making the data into more of a normal distribution. This change makes it more useful for modeling purposes so the Box-Cox’d version of Serum Cholesterol will be used from here on out.
Including the details above, this step also involved:
- Statistical Analysis
- Normality Tests
- Chi-Square Tests
- Contingency Tables
- Odds Ratios
- Descriptive Statistics
- Additional Feature Engineering
- Additional Data Visualizations
- Additional Data Transformations
Model Building (View code)
After exploring our data to obtain a greater understanding of it and using that information to perform feature engineering and data transformations, it was time to build and optimize models.
- Five different machine learning algorithms were used:
- Logistic Regression
- Random Forest
- K-Nearest Neighbors
- Support Vector Machine
- Gradient Boosting
- Seven unique sets of variables were created with each set containing continuous and categorical features
- Each model was run on every set of variables for a total of 35 models
- Every model run was optimized using:
- Grid search
- Cross-validation
- Feature importance techniques
Model Visualization, Comparison, and Selection (View code)
- ROC Curves were built based on each model’s predicted probabilities to visually compare model performance at various cut-off values.
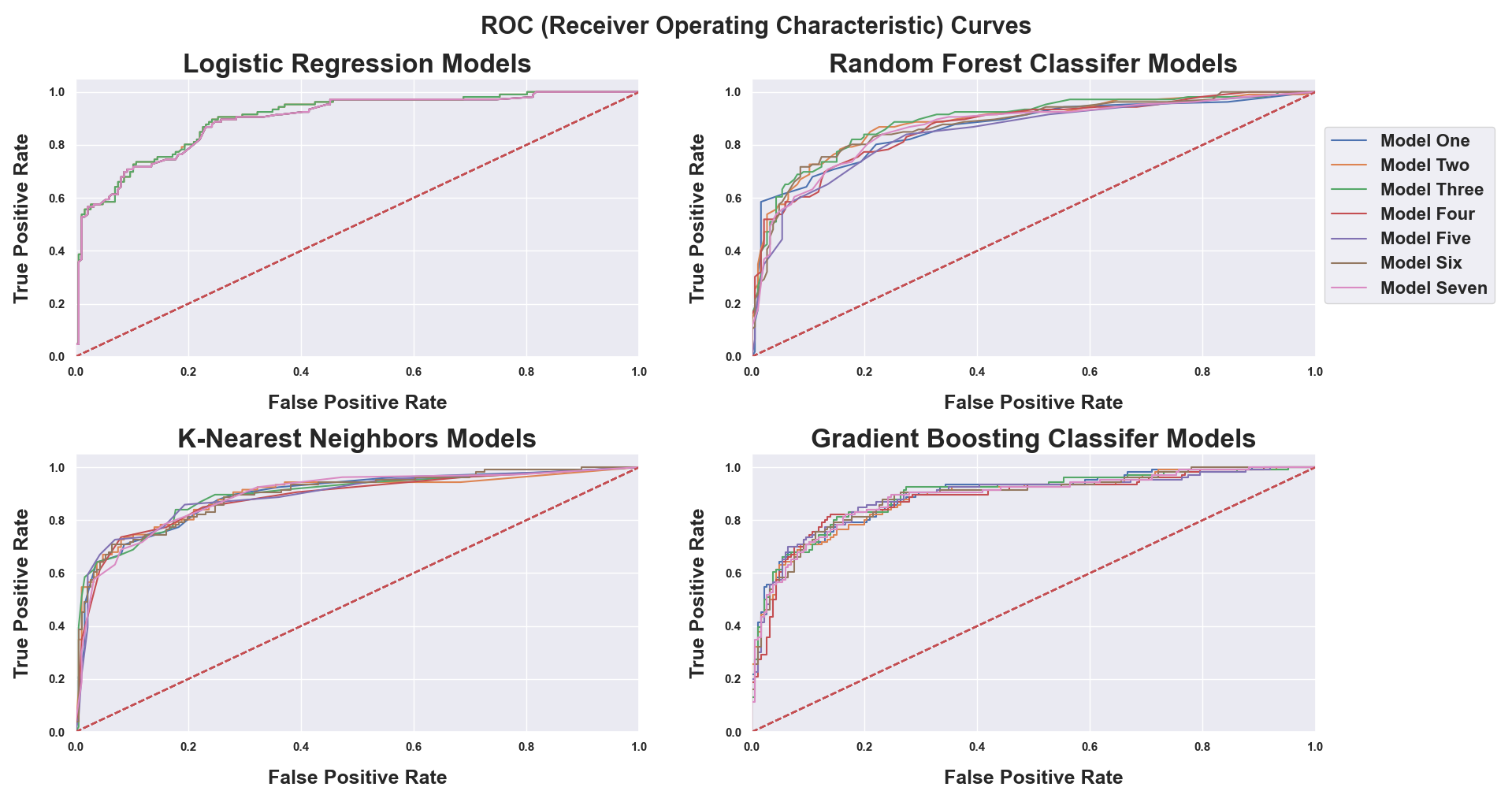
The four models that give predicted probabilities (Support Vector Machines do not give predicted probabilities, only class membership) are plotted above, and each plot contains seven ROC curves - one for each unique model run. The most amount of variation can be seen in the Random Forest Classifier models, and the least amount in the Logistic Regression models due to the fact variables had to be statistically significant to be included in the latter model.
The best model for each of the five algorithms was selected based on F1 Score. For the four models that gave predicted probabilities, the cut-off value chosen for each model was the one that provided the optimal combination of total patients predicted correctly and F1 Score. From there, model predictions were assembled to determine which combination (or stand alone model) provided the best results.
- A summary of the model results can be seen below.
- The best model, the Support Vector Machine Classification Model #4, is bolded. It is the most parsimonious model that provided the optimal combination of total patients predicted correctly and F1 Score.
| Model(s) | F1 Score | Recall | Precision | Total Correct | Total Incorrect |
|---|---|---|---|---|---|
| Gradient Boosting Classifier Five, K-Nearest Neighbors Five, Support Vector Machine Classifier Four | 0.802 | 0.745 | 0.868 | 253 | 39 |
| Support Vector Machine Classifier Four | 0.804 | 0.774 | 0.837 | 252 | 40 |
| Gradient Boosting Classifier Five, Random Forest Classifier Six, Support Vector Machine Classifier Four | 0.798 | 0.745 | 0.859 | 252 | 40 |
| Gradient Boosting Classifier Five, K-Nearest Neighbors Five, Logistic Regression Three, Random Forest Classifier Six, Support Vector Machine Classifier Four | 0.798 | 0.745 | 0.859 | 252 | 40 |
| Logistic Regression Three, Random Forest Classifier Six, Support Vector Machine Classifier Four | 0.792 | 0.755 | 0.833 | 250 | 42 |
| K-Nearest Neighbors Five | 0.786 | 0.726 | 0.856 | 250 | 42 |
| K-Nearest Neighbors Five, Random Forest Classifier Six, Support Vector Machine Classifier Four | 0.786 | 0.726 | 0.856 | 250 | 42 |
| K-Nearest Neighbors Five, Logistic Regression Three, Support Vector Machine Classifier Four | 0.786 | 0.745 | 0.832 | 249 | 43 |
| Gradient Boosting Classifier Five, K-Nearest Neighbors Five, Logistic Regression Three | 0.782 | 0.726 | 0.846 | 249 | 43 |
| Gradient Boosting Classifier Five, Logistic Regression Three, Support Vector Machine Classifier Four | 0.782 | 0.726 | 0.846 | 249 | 43 |
| Gradient Boosting Classifier Five | 0.771 | 0.698 | 0.860 | 248 | 44 |
| Gradient Boosting Classifier Five, Logistic Regression Three, Random Forest Classifier Six | 0.772 | 0.717 | 0.835 | 247 | 45 |
| K-Nearest Neighbors Five, Logistic Regression Three, Random Forest Classifier Six | 0.772 | 0.717 | 0.835 | 247 | 45 |
| Gradient Boosting Classifier Five, K-Nearest Neighbors Five, Random Forest Classifier Six | 0.769 | 0.708 | 0.843 | 247 | 45 |
| Random Forest Classifier Six | 0.768 | 0.717 | 0.826 | 246 | 46 |
| Logistic Regression Three | 0.765 | 0.736 | 0.796 | 244 | 48 |
Visualize Best Model (View code)
The next step was visualizing how the best model performed in an easy to understand way.
The first visual below - a confusion matrix - is a fundamental assessment tool for classification problems. It is a cross tabulation of the actual and predicted classes and quantifies the confusion of the classifier. Here it details the prediction results of the best model, the Support Vector Machine Classification Model #4.
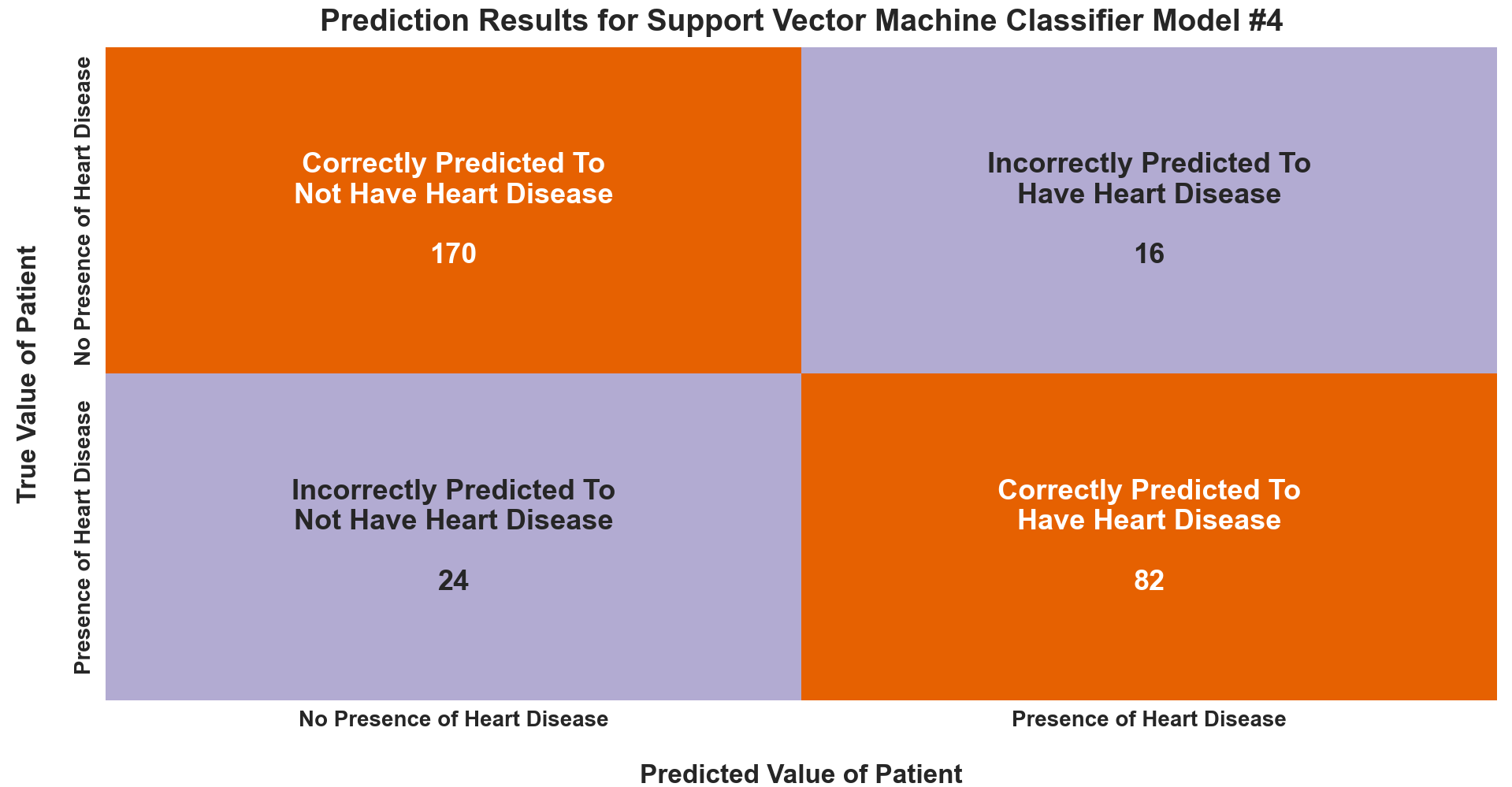
- The top left corner of the confusion matrix indicates the 170 patients who had no presence of heart disease (i.e., their actual value) and were predicted as such (i.e., their predicted value). These are labeled as true negatives.
- The top right corner, designated as false positives, denotes the 16 patients who had no presence of heart disease but were deemed to have heart disease by the model.
- The bottom left corner, known as false negatives, represents the 24 patients who actually had the presence of heart disease but were predicted by the model to not have heart disease.
- The bottom right corner shows the 82 patients who had a presence of heart disease and were correctly predicted by the model to have that presence. These patients are signified as true positives.
When looking at a confusion matrix, we want the true negative and true positive values to be high and the false positive and false negative values to be low. This indicates a highly accurate model, which is exactly what we have here.
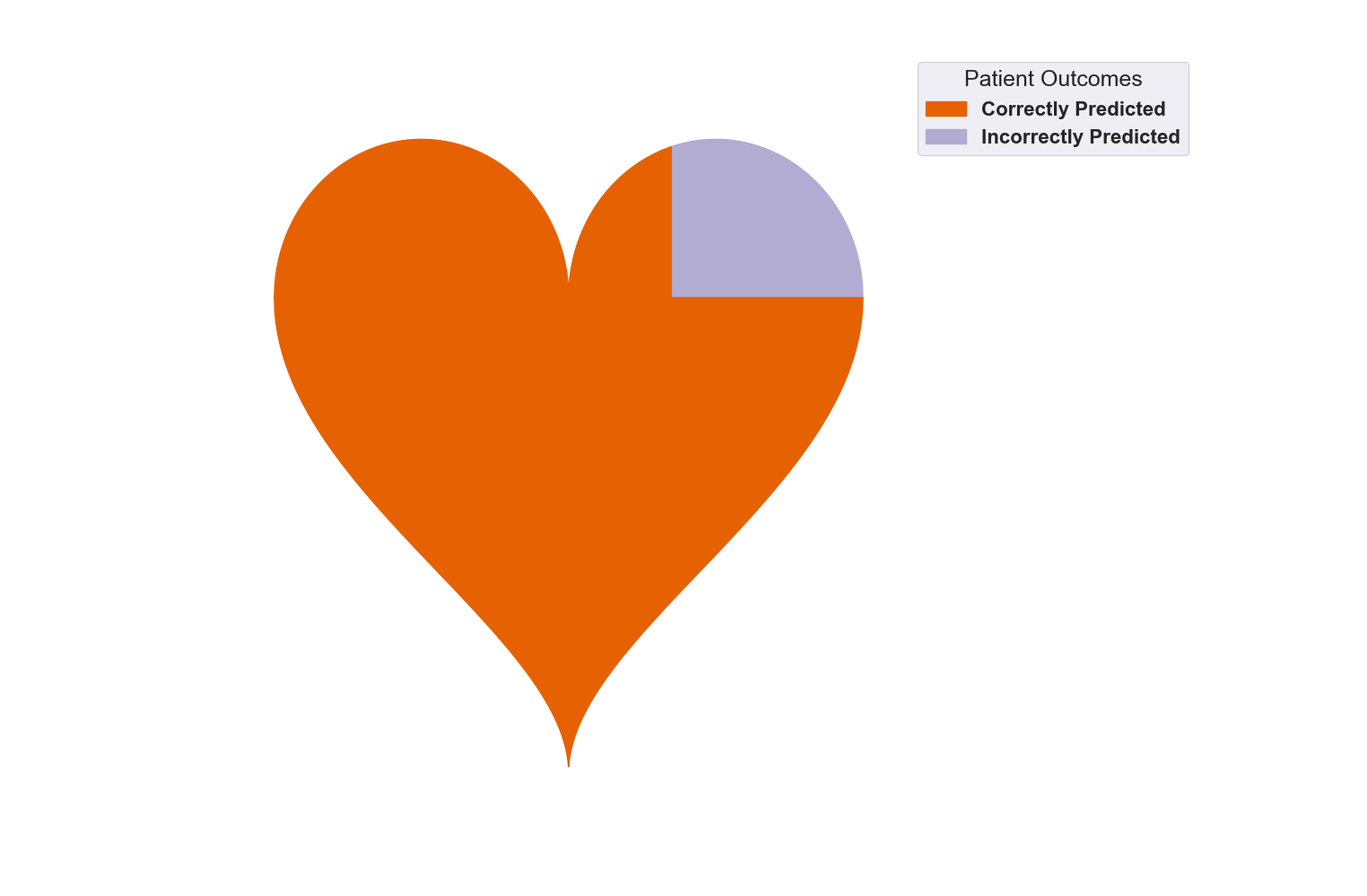
The next visualization shows the same results as above, just now in a more concise format. This stacked bar chart condenses our results into patients who were correctly predicted and those who were incorrectly predicted.
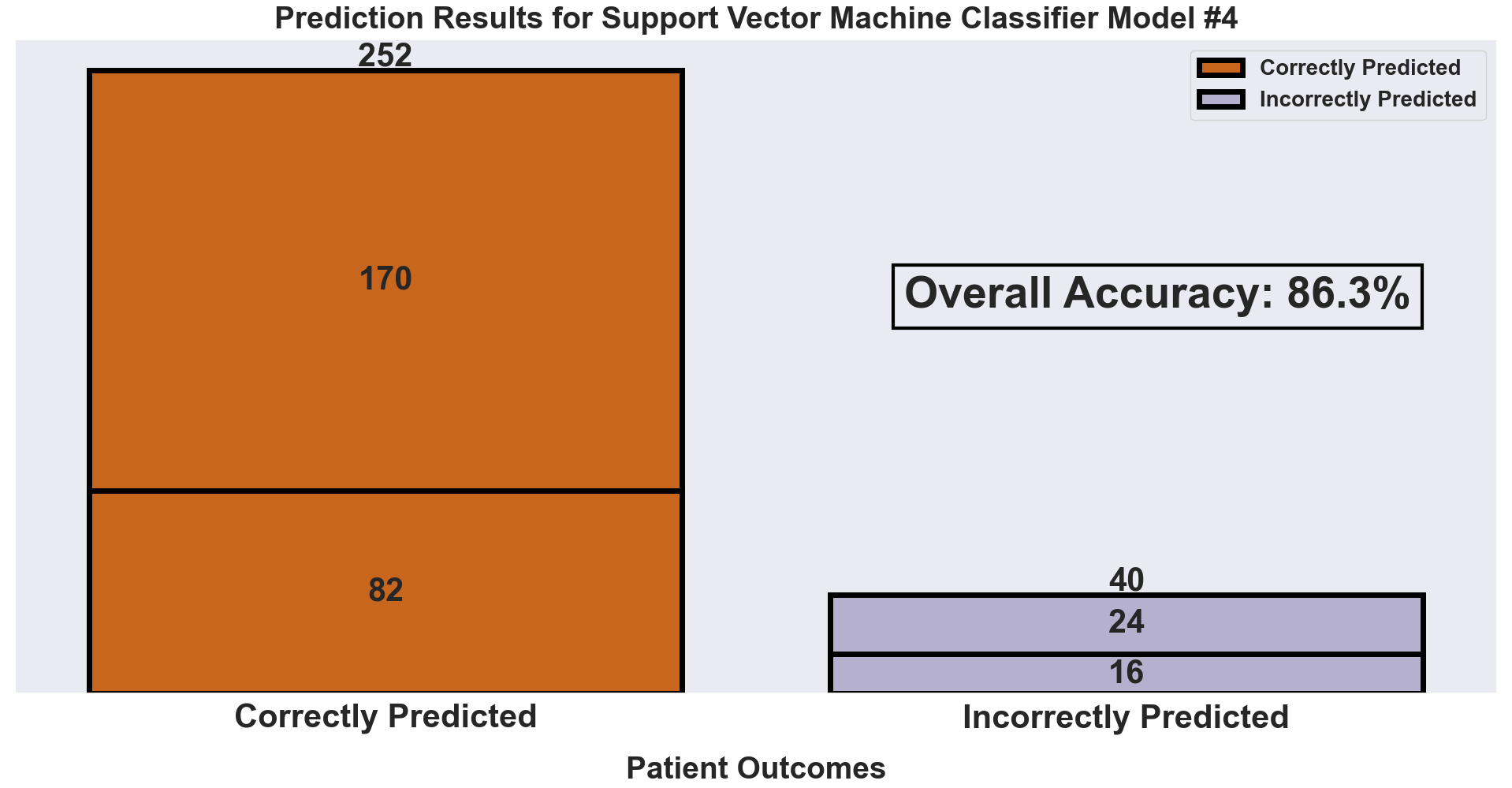
- As expected, the true negatives and true positives make up our ‘Correctly Predicted’ bar while the false positives and false negatives make up our ‘Incorrectly Predicted’ bar.
- It is easy to see how successful the model is by how much larger the ‘Correctly Predicted’ bar is than the ‘Incorrectly Predicted’ bar. The model was able to accurately predict 252 patients while only missing 40 patients.
Model Usefulness
The final step, and arguably the most critical one, is explaining how the results could be utilized in a medical facility setting to benefit medical practitioners and their patients.
- The model could be implemented, along with the patient’s formal checkups and examinations, to assist medical practitioners in correctly diagnosing heart disease in their patients.
- It would allow practitioners to get an in-depth understanding of which factors contribute to heart disease, and set up a prehabilitation routine for their patients that would help decrease those factors.
- Parts of the routine would include helping the patient establish a diet to decrease their serum cholesterol and formulating an exercise regimen that could help lower their blood pressure.
- This would provide patients a path towards a clean bill of health, and prevent possible heart disease in the future.
- With the patient being in a better state of health, they can avoid distressing tests and procedures, evade medications, and get back to leading a productive, wholesome life.
Questions for Consideration
- How else could the model be useful?
- What are other visualizations that could be useful to understand the data and results?
- Are there external variables or ones that could be engineered from the existing set that could potentially improve the model’s performance?
Data Ingestion
Import Libraries and Packages
import os
import yaml
import pickle
import numpy as np
import pandas as pd
import datetime as dt
from math import sqrt
from scipy import stats
import statsmodels.api as sm
import inflect
import itertools
from more_itertools import unique_everseen
from sklearn.decomposition import PCA
from sklearn.feature_selection import RFE
from sklearn.preprocessing import StandardScaler
from sklearn.svm import SVC
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier, KNeighborsRegressor
from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier
from sklearn.metrics import confusion_matrix, roc_curve
from sklearn.model_selection import cross_val_score, cross_val_predict, train_test_split, GridSearchCV, ShuffleSplit
from sklearn.exceptions import ConvergenceWarning
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
import seaborn as sns
# Increase maximum width in characters of columns - will put all columns in same line in console readout
pd.set_option('expand_frame_repr', False)
# Be able to read entire value in each column (no longer truncating values)
pd.set_option('display.max_colwidth', -1)
# Increase number of rows printed out in console
pd.set_option('display.max_rows', 200)
# Set aesthetic parameters of seaborn plots
sns.set()
# Change current working directory to main directory
def main_directory():
# Load in .yml file to retrieve location of heart disease directory
info = yaml.load(open("info.yml"), Loader=yaml.FullLoader)
os.chdir(os.getcwd() + info['heart_disease_directory'])
main_directory()
# Open Hungarian data set
with open('hungarian.data', 'r') as myfile:
file = []
for line in myfile:
line = line.replace(" ", ", ")
# Add comma to end of each line
line = line.replace(os.linesep, ',' + os.linesep)
line = line.split(', ')
file.extend(line)
file = [value.replace(",\n", "") for value in file]
# Remove empty strings from list
file = list(filter(None, file))
# Convert list to lists of list
attributes_per_patient = int(len(file)/294) # len(file)/number of patients
i = 0
new_file = []
while i < len(file):
new_file.append(file[i:i+attributes_per_patient])
i += attributes_per_patient
# List of column names
headers = ['id', 'ccf', 'age', 'sex', 'painloc', 'painexer', 'relrest', 'pncaden', 'cp', 'trestbps', 'htn', 'chol',
'smoke', 'cigs', 'years', 'fbs', 'dm', 'famhist', 'restecg', 'ekgmo', 'ekgday', 'ekgyr', 'dig', 'prop',
'nitr', 'pro', 'diuretic', 'proto', 'thaldur', 'thaltime', 'met', 'thalach', 'thalrest', 'tpeakbps',
'tpeakbpd', 'dummy', 'trestbpd', 'exang', 'xhypo', 'oldpeak', 'slope', 'rldv5', 'rldv5e', 'ca', 'restckm',
'exerckm', 'restef', 'restwm', 'exeref', 'exerwm', 'thal', 'thalsev', 'thalpul', 'earlobe', 'cmo',
'cday', 'cyr', 'num', 'lmt', 'ladprox', 'laddist', 'diag', 'cxmain', 'ramus', 'om1', 'om2', 'rcaprox',
'rcadist', 'lvx1', 'lvx2', 'lvx3', 'lvx4', 'lvf', 'cathef', 'junk', 'name']
# Convert lists of list into DataFrame and supply column names
hungarian = pd.DataFrame(new_file, columns=headers)
Data Cleaning
Remove Unnecessary Columns
# List of columns to drop
cols_to_drop =['ccf', 'pncaden', 'smoke', 'cigs', 'years', 'dm', 'famhist', 'dig', 'ca', 'restckm', 'exerckm',
'restef', 'restwm', 'exeref', 'exerwm', 'thal', 'thalsev', 'thalpul', 'earlobe', 'lmt',
'ladprox', 'laddist', 'diag', 'cxmain', 'ramus', 'om1', 'om2', 'rcaprox', 'rcadist', 'lvx1',
'cathef', 'junk', 'name', 'thaltime', 'xhypo', 'slope', 'dummy', 'lvx1', 'lvx2']
# Drop columns from above list
hungarian = hungarian.drop(columns=cols_to_drop)
Convert Column Types
# Convert all columns to numeric
hungarian = hungarian.apply(pd.to_numeric)
Correct Data Discrepancies
### Fix possible patient id issues
# Find ids that are not unique to patients
print(hungarian.id.value_counts()[hungarian.id.value_counts()!=1])
# Fix id 1132 (two different patients are both assigned to this id) - give second patient next id number (id max + 1)
hungarian.loc[hungarian.loc[hungarian.id==1132].index[-1], 'id'] = hungarian.id.max() + 1
Remove Patients with a Large Percentage of Missing Values
# Drop patients with "significant" number of missing values (use 10%, can adjust accordingly)
# Determine missing value percentage per patient (-9 is the missing attribute value)
missing_value_perc_per_patient = (hungarian == -9).sum(axis=1)[(hungarian == -9).sum(axis=1) > 0]\
.sort_values(ascending=False)/len([x for x in hungarian.columns if x != 'id'])
# Remove patients with > 10% missing values
hungarian = hungarian.drop(missing_value_perc_per_patient[missing_value_perc_per_patient>0.10].index.values)
Impute Missing Values Using K-Nearest Neighbors
### Imputing missing values (marked as -9 per data dictionary)
cols_with_missing_values = [(col, hungarian[col].value_counts()[-9]) for col in list(hungarian) if -9 in hungarian[col].unique()]
# Sort tuples by number of missing values
cols_with_missing_values.sort(key=lambda x: x[1])
# Use K-Nearest Neighbors (KNN) to impute missing values
# Method to scale continuous and binary variables (z-score standardization)
scaler = StandardScaler()
variables_not_to_use_for_imputation = ['ekgday', 'cmo', 'cyr', 'ekgyr', 'cday', 'ekgmo', 'num']
# Impute htn
impute_variable = 'htn'
# Obtain list of variables to use for imputation
x_variables = [x for x in list(hungarian) if x not in [x[0] for x in cols_with_missing_values] +
variables_not_to_use_for_imputation + ['id']]
# Select x and y variables to use
fix_htn = hungarian[x_variables + [impute_variable]]
# Create list of categorical variables to one-hot encode
categorical_x_variables = ['sex', 'painloc', 'painexer', 'relrest', 'cp', 'nitr', 'pro', 'diuretic', 'exang',
'lvx3', 'lvx4', 'lvf']
# One-hot encode categorical variables
for value in categorical_x_variables:
one_hot = pd.get_dummies(fix_htn[value], prefix=value)
fix_htn = fix_htn.join(one_hot)
fix_htn = fix_htn.drop(columns=value)
# Create list of x variables
x_variables = [x for x in list(fix_htn) if x != impute_variable]
# Create DataFrame with missing value(s) to predict on
predict = fix_htn.loc[fix_htn[impute_variable]==-9]
# Set x and y predict DataFrames
predict_x, predict_y = predict[x_variables], predict[impute_variable]
# Create DataFrame to train on
train = fix_htn.loc[~(fix_htn[impute_variable]==-9)]
# Set x and y train DataFrames
train_x, train_y = train[x_variables], train[impute_variable]
# Fit and transform scaler on train_x
train_x = scaler.fit_transform(train_x)
# Transform predict_x
predict_x = scaler.transform(predict_x)
# Obtain k (number of neighbors) by using sqrt(n)
k = round(sqrt(len(train_x)))
print(f"k is {k}.")
# Check to make sure k is odd number
if divmod(k, 2)[1] == 1:
print("k is an odd number. Good to proceed.")
else:
print("Need to make k an odd number.")
# Predict value for predict_y
htn_prediction = KNeighborsClassifier(n_neighbors=k, metric='minkowski', weights='distance').fit(train_x, train_y).predict(predict_x)
print(f'The prediction for htn is {htn_prediction[0]}.')
# Supply prediction back to appropriate patient
hungarian.loc[hungarian[impute_variable]==-9, 'htn'] = htn_prediction
# Imputing missing values (marked as -9 per data dictionary)
cols_with_missing_values = [(col, hungarian[col].value_counts()[-9]) for col in list(hungarian) if -9 in hungarian[col].unique()]
# Sort tuples by number of missing values
cols_with_missing_values.sort(key=lambda x:x[1])
# Impute restecg
impute_variable = 'restecg'
# Obtain list of variables to use for imputation
x_variables = [x for x in list(hungarian) if x not in [x[0] for x in cols_with_missing_values] +
variables_not_to_use_for_imputation + ['id']]
# Select x and y variables to use - added in 'htn'
fix_restecg = hungarian[x_variables + [impute_variable]]
# Create list of categorical variables to one-hot encode
categorical_x_variables = ['sex', 'painloc', 'painexer', 'relrest', 'cp', 'htn', 'nitr', 'pro', 'diuretic', 'exang',
'lvx3', 'lvx4', 'lvf']
# One-hot encode categorical variables
for value in categorical_x_variables:
one_hot = pd.get_dummies(fix_restecg[value], prefix=value)
fix_restecg = fix_restecg.join(one_hot)
fix_restecg = fix_restecg.drop(columns=value)
# Create list of x variables
x_variables = [x for x in list(fix_restecg) if x != impute_variable]
# Create DataFrame with missing value(s) - will predict on
predict = fix_restecg.loc[fix_restecg[impute_variable]==-9]
# Set x and y predict DataFrames
predict_x, predict_y = predict[x_variables], predict[impute_variable]
# Create DataFrame to train on
train = fix_restecg.loc[~(fix_restecg[impute_variable]==-9)]
# Set x and y train DataFrames
train_x, train_y = train[x_variables], train[impute_variable]
# Fit and transform scaler on train_x
train_x = scaler.fit_transform(train_x)
# Transform predict_x
predict_x = scaler.transform(predict_x)
# Obtain k (number of neighbors) by using sqrt(n)
k = round(sqrt(len(train_x)))
print(f"k is {k}.")
# Check to make sure k is odd number
if divmod(k, 2)[1] == 1:
print("k is an odd number. Good to proceed.")
else:
print("Need to make k an odd number.")
# Predict value for predict_y
restecg_prediction = KNeighborsClassifier(n_neighbors=k, metric='minkowski', weights='distance').fit(train_x, train_y).predict(predict_x)
print(f'The prediction for restecg is {restecg_prediction[0]}.')
# Supply prediction back to appropriate patient
hungarian.loc[hungarian[impute_variable]==-9, 'restecg'] = restecg_prediction
# Imputing missing values (marked as -9 per data dictionary)
cols_with_missing_values = [(col, hungarian[col].value_counts()[-9]) for col in list(hungarian) if -9 in hungarian[col].unique()]
# Sort tuples by number of missing values
cols_with_missing_values.sort(key=lambda x:x[1])
# Impute prop
# Set y variable
impute_variable = 'prop'
# Obtain list of variables to use for imputation
x_variables = [x for x in list(hungarian) if x not in [x[0] for x in cols_with_missing_values] +
variables_not_to_use_for_imputation + ['id']]
# Select x and y variables to use - add in 'htn'
fix_prop = hungarian[x_variables + [impute_variable]]
# Create list of categorical variables to one-hot encode
categorical_x_variables = ['sex', 'painloc', 'painexer', 'relrest', 'cp', 'htn', 'restecg', 'nitr', 'pro', 'diuretic', 'exang',
'lvx3', 'lvx4', 'lvf']
# One-hot encode categorical variables
for value in categorical_x_variables:
one_hot = pd.get_dummies(fix_prop[value], prefix=value)
fix_prop = fix_prop.join(one_hot)
fix_prop = fix_prop.drop(columns=value)
# Create list of x variables
x_variables = [x for x in list(fix_prop) if x != impute_variable]
# Create DataFrame with missing value(s) - will predict on
predict = fix_prop.loc[fix_prop[impute_variable]==-9]
# Set x and y predict DataFrames
predict_x, predict_y = predict[x_variables], predict[impute_variable]
# Create DataFrame to train on
train = fix_prop.loc[~(fix_prop[impute_variable]==-9)]
# Set x and y train DataFrames
train_x, train_y = train[x_variables], train[impute_variable]
# Fit and transform scaler on train_x
train_x = scaler.fit_transform(train_x)
# Transform predict_x
predict_x = scaler.transform(predict_x)
# Obtain k (number of neighbors) by using sqrt(n)
k = round(sqrt(len(train_x)))
print(f"k is {k}.")
# Check to make sure k is odd number
if divmod(k, 2)[1] == 1:
print("k is an odd number. Good to proceed.")
else:
print("Need to make k an odd number.")
# Predict value for predict_y
prop_prediction = KNeighborsClassifier(n_neighbors=k, metric='minkowski', weights='distance').fit(train_x, train_y).predict(predict_x)
print(f'The prediction for prop is {prop_prediction[0]}.')
# Supply prediction back to appropriate patient
hungarian.loc[hungarian[impute_variable]==-9, 'prop'] = prop_prediction
# Imputing missing values (marked as -9 per data dictionary)
cols_with_missing_values = [(col, hungarian[col].value_counts()[-9]) for col in list(hungarian) if -9 in hungarian[col].unique()]
# Sort tuples by number of missing values
cols_with_missing_values.sort(key=lambda x:x[1])
# Impute thaldur
# Set y variable
impute_variable = 'thaldur'
# Obtain list of variables to use for imputation
x_variables = [x for x in list(hungarian) if x not in [x[0] for x in cols_with_missing_values] +
variables_not_to_use_for_imputation + ['id']]
# Select x and y variables to use - add in 'prop'
fix_thaldur = hungarian[x_variables + [impute_variable]]
# Create list of categorical variables to one-hot encode
categorical_x_variables = ['sex', 'painloc', 'painexer', 'relrest', 'cp', 'htn', 'restecg', 'prop', 'nitr', 'pro',
'diuretic', 'exang', 'lvx3', 'lvx4', 'lvf']
# One-hot encode categorical variables
for value in categorical_x_variables:
one_hot = pd.get_dummies(fix_thaldur[value], prefix=value)
fix_thaldur = fix_thaldur.join(one_hot)
fix_thaldur = fix_thaldur.drop(columns=value)
# Create list of x variables
x_variables = [x for x in list(fix_thaldur) if x != impute_variable]
# Create DataFrame with missing value(s) - will predict on
predict = fix_thaldur.loc[fix_thaldur[impute_variable]==-9]
# Set x and y predict DataFrames
predict_x, predict_y = predict[x_variables], predict[impute_variable]
# Create DataFrame to train on
train = fix_thaldur.loc[~(fix_thaldur[impute_variable]==-9)]
# Set x and y train DataFrames
train_x, train_y = train[x_variables], train[impute_variable]
# Fit scaler on train_x
train_x = scaler.fit_transform(train_x)
# Transform predict_x
predict_x = scaler.transform(predict_x)
# Obtain k (number of neighbors) by using sqrt(n)
k = round(sqrt(len(train_x)))
print(f"k is {k}.")
# Check to make sure k is odd number
if divmod(k, 2)[1] == 1:
print("k is an odd number. Good to proceed.")
else:
print("Need to make k an odd number.")
# Predict value for predict_y
thaldur_prediction = KNeighborsRegressor(n_neighbors=k, metric='minkowski', weights='distance').fit(train_x, train_y).predict(predict_x)
print("The prediction for thaldur is " + str(thaldur_prediction[0]) + ".")
# Round thaldur_prediction to integer
thaldur_prediction = round(number=thaldur_prediction[0])
print("The prediction for thaldur has been rounded to " + str(thaldur_prediction) + ".")
# Supply prediction back to appropriate patient
hungarian.loc[hungarian[impute_variable]==-9, 'thaldur'] = thaldur_prediction
# Imputing missing values (marked as -9 per data dictionary)
cols_with_missing_values = [(col, hungarian[col].value_counts()[-9]) for col in list(hungarian) if -9 in hungarian[col].unique()]
# Sort tuples by number of missing values
cols_with_missing_values.sort(key=lambda x:x[1])
# Impute rldv5
# Set y variable
impute_variable = 'rldv5'
# Obtain list of variables to use for imputation
x_variables = [x for x in list(hungarian) if x not in [x[0] for x in cols_with_missing_values] +
variables_not_to_use_for_imputation + ['id']]
# Select x and y variables to use - add in 'prop'
fix_rldv5 = hungarian[x_variables + [impute_variable]]
# Create list of categorical variables to one-hot encode
categorical_x_variables = ['sex', 'painloc', 'painexer', 'relrest', 'cp', 'htn', 'restecg', 'prop', 'nitr', 'pro',
'diuretic', 'exang', 'lvx3', 'lvx4', 'lvf']
# One-hot encode categorical variables
for value in categorical_x_variables:
one_hot = pd.get_dummies(fix_rldv5[value], prefix=value)
fix_rldv5 = fix_rldv5.join(one_hot)
fix_rldv5 = fix_rldv5.drop(columns=value)
# Create list of x variables
x_variables = [x for x in list(fix_rldv5) if x != impute_variable]
# Create DataFrame with missing value(s) - will predict on
predict = fix_rldv5.loc[fix_rldv5[impute_variable]==-9]
# Set x and y predict DataFrames
predict_x, predict_y = predict[x_variables], predict[impute_variable]
# Create DataFrame to train on
train = fix_rldv5.loc[~(fix_rldv5[impute_variable]==-9)]
# Set x and y train DataFrames
train_x, train_y = train[x_variables], train[impute_variable]
# Fit scaler on train_x
train_x = scaler.fit_transform(train_x)
# Transform predict_x
predict_x = scaler.transform(predict_x)
# Obtain k (number of neighbors) by using sqrt(n)
k = round(sqrt(len(train_x)))
print(f"k is {k}.")
# Check to make sure k is odd number
if divmod(k, 2)[1] == 1:
print("k is an odd number. Good to proceed.")
else:
print("Need to make k an odd number.")
# Predict value for predict_y
rldv5_prediction = KNeighborsRegressor(n_neighbors=k, metric='minkowski', weights='distance').fit(train_x, train_y).predict(predict_x)
print("The prediction for rldv5 is " + str(rldv5_prediction[0]) + ".")
# Round rldv5_prediction to integer
rldv5_prediction = round(number=rldv5_prediction[0])
print("The prediction for rldv5 has been rounded to " + str(rldv5_prediction) + ".")
# Supply prediction back to appropriate patient
hungarian.loc[hungarian[impute_variable]==-9, 'rldv5'] = rldv5_prediction
# Imputing missing values (marked as -9 per data dictionary)
cols_with_missing_values = [(col, hungarian[col].value_counts()[-9]) for col in list(hungarian) if -9 in hungarian[col].unique()]
# Sort tuples by number of missing values
cols_with_missing_values.sort(key=lambda x:x[1])
# Impute met
# Set y variable
impute_variable = 'met'
# Obtain list of variables to use for imputation
x_variables = [x for x in list(hungarian) if x not in [x[0] for x in cols_with_missing_values] +
variables_not_to_use_for_imputation + ['id']]
# Select x and y variables to use - add in 'rldv5'
fix_met = hungarian[x_variables + [impute_variable]]
# Create list of categorical variables to one-hot encode
categorical_x_variables = ['sex', 'painloc', 'painexer', 'relrest', 'cp', 'htn', 'restecg', 'prop', 'nitr', 'pro',
'diuretic', 'exang', 'lvx3', 'lvx4', 'lvf']
# One-hot encode categorical variables
for value in categorical_x_variables:
one_hot = pd.get_dummies(fix_met[value], prefix=value)
fix_met = fix_met.join(one_hot)
fix_met = fix_met.drop(columns=value)
# Create list of x variables
x_variables = [x for x in list(fix_met) if x != impute_variable]
# Create DataFrame with missing value(s) - will predict on
predict = fix_met.loc[fix_met[impute_variable]==-9]
# Set x and y predict DataFrames
predict_x, predict_y = predict[x_variables], predict[impute_variable]
# Create DataFrame to train on
train = fix_met.loc[~(fix_met[impute_variable]==-9)]
# Set x and y train DataFrames
train_x, train_y = train[x_variables], train[impute_variable]
# Fit scaler on train_x
train_x = scaler.fit_transform(train_x)
# Transform predict_x
predict_x = scaler.transform(predict_x)
# Obtain k (number of neighbors) by using sqrt(n)
k = round(sqrt(len(train_x)))
print(f"k is {k}.")
# Check to make sure k is odd number
if divmod(k, 2)[1] == 1:
print("k is an odd number. Good to proceed.")
else:
print("Need to make k an odd number.")
# Predict value for predict_y
met_prediction = KNeighborsRegressor(n_neighbors=k, metric='minkowski', weights='distance').fit(train_x, train_y).predict(predict_x)
print("The predictions for met are:")
print(met_prediction)
# Round met_prediction to integer
for i in range(len(met_prediction)):
met_prediction[i] = round(number=met_prediction[i])
print("The prediction for met_prediction" + "[" + str(i) + "]" + " has been rounded to " + str(met_prediction[i]) + ".")
# Supply prediction back to appropriate patient
hungarian.loc[hungarian[impute_variable]==-9, impute_variable] = met_prediction
# Imputing missing values (marked as -9 per data dictionary)
cols_with_missing_values = [(col, hungarian[col].value_counts()[-9]) for col in list(hungarian) if -9 in hungarian[col].unique()]
# Sort tuples by number of missing values
cols_with_missing_values.sort(key=lambda x:x[1])
# Impute fbs
# Set y variable
impute_variable = 'fbs'
# Obtain list of variables to use for imputation
x_variables = [x for x in list(hungarian) if x not in [x[0] for x in cols_with_missing_values] +
variables_not_to_use_for_imputation + ['id']]
# Select x and y variables to use - add in 'met'
fix_fbs = hungarian[x_variables + [impute_variable]]
# Create list of categorical variables to one-hot encode
categorical_x_variables = ['sex', 'painloc', 'painexer', 'relrest', 'cp', 'htn', 'restecg', 'prop', 'nitr', 'pro',
'diuretic', 'exang', 'lvx3', 'lvx4', 'lvf']
# One-hot encode categorical variables
for value in categorical_x_variables:
one_hot = pd.get_dummies(fix_fbs[value], prefix=value)
fix_fbs = fix_fbs.join(one_hot)
fix_fbs = fix_fbs.drop(columns=value)
# Create list of x variables
x_variables = [x for x in list(fix_fbs) if x != impute_variable]
# Create DataFrame with missing value(s) - will predict on
predict = fix_fbs.loc[fix_fbs[impute_variable]==-9]
# Set x and y predict DataFrames
predict_x, predict_y = predict[x_variables], predict[impute_variable]
# Create DataFrame to train on
train = fix_fbs.loc[~(fix_fbs[impute_variable]==-9)]
# Set x and y train DataFrames
train_x, train_y = train[x_variables], train[impute_variable]
# Fit scaler on train_x
train_x = scaler.fit_transform(train_x)
# Transform predict_x
predict_x = scaler.transform(predict_x)
# Obtain k (number of neighbors) by using sqrt(n)
k = round(sqrt(len(train_x)))
print(f"k is {k}.")
# Check to make sure k is odd number
if divmod(k, 2)[1] == 1:
print("k is an odd number. Good to proceed.")
else:
print("Need to make k an odd number.")
# Predict value for predict_y
fbs_prediction = KNeighborsClassifier(n_neighbors=k, metric='minkowski', weights='distance').fit(train_x, train_y).predict(predict_x)
print("The predictions for fbs are:")
print(fbs_prediction)
# Supply prediction back to appropriate patient
hungarian.loc[hungarian[impute_variable]==-9, impute_variable] = fbs_prediction
# Imputing missing values (marked as -9 per data dictionary)
cols_with_missing_values = [(col, hungarian[col].value_counts()[-9]) for col in list(hungarian) if -9 in hungarian[col].unique()]
# Sort tuples by number of missing values
cols_with_missing_values.sort(key=lambda x:x[1])
# Impute fbs
# Set y variable
impute_variable = 'proto'
# Obtain list of variables to use for imputation
x_variables = [x for x in list(hungarian) if x not in [x[0] for x in cols_with_missing_values] +
variables_not_to_use_for_imputation + ['id']]
# Select x and y variables to use - add in 'fbs'
fix_proto = hungarian[x_variables + [impute_variable]]
# Create list of categorical variables to one-hot encode
categorical_x_variables = ['sex', 'painloc', 'painexer', 'relrest', 'cp', 'htn', 'fbs', 'restecg', 'prop', 'nitr', 'pro',
'diuretic', 'exang', 'lvx3', 'lvx4', 'lvf']
# One-hot encode categorical variables
for value in categorical_x_variables:
one_hot = pd.get_dummies(fix_proto[value], prefix=value)
fix_proto = fix_proto.join(one_hot)
fix_proto = fix_proto.drop(columns=value)
# Create list of x variables
x_variables = [x for x in list(fix_proto) if x != impute_variable]
# Create DataFrame with missing value(s) - will predict on
predict = fix_proto.loc[fix_proto[impute_variable]==-9]
# Set x and y predict DataFrames
predict_x, predict_y = predict[x_variables], predict[impute_variable]
# Create DataFrame to train on
train = fix_proto.loc[~(fix_proto[impute_variable]==-9)]
# Set x and y train DataFrames
train_x, train_y = train[x_variables], train[impute_variable]
# Fit scaler on train_x
train_x = scaler.fit_transform(train_x)
# Transform train_x
train_x = scaler.transform(train_x)
# Transform predict_x
predict_x = scaler.transform(predict_x)
# Obtain k (number of neighbors) by using sqrt(n)
k = round(sqrt(len(train_x)))
print(f"k is {k}.")
# Check to make sure k is odd number
if divmod(k, 2)[1] == 1:
print("k is an odd number. Good to proceed.")
else:
print("Need to make k an odd number.")
# Predict value for predict_y
proto_prediction = KNeighborsClassifier(n_neighbors=k, metric='minkowski', weights='distance').fit(train_x, train_y).predict(predict_x)
print("The predictions for proto are:")
print(proto_prediction)
# Supply prediction back to appropriate patient
hungarian.loc[hungarian[impute_variable]==-9, impute_variable] = proto_prediction
# Imputing missing values (marked as -9 per data dictionary)
cols_with_missing_values = [(col, hungarian[col].value_counts()[-9]) for col in list(hungarian) if -9 in hungarian[col].unique()]
# Sort tuples by number of missing values
cols_with_missing_values.sort(key=lambda x:x[1])
# Impute chol
impute_variable = 'chol'
# Obtain list of variables to use for imputation
x_variables = [x for x in list(hungarian) if x not in [x[0] for x in cols_with_missing_values] +
variables_not_to_use_for_imputation + ['id']]
# Select x and y variables to use - add in 'fbs'
fix_chol = hungarian[x_variables + [impute_variable]]
# Create list of categorical variables to one-hot encode
categorical_x_variables = ['sex', 'painloc', 'painexer', 'relrest', 'cp', 'htn', 'fbs', 'restecg', 'prop', 'nitr', 'pro',
'diuretic', 'proto', 'exang', 'lvx3', 'lvx4', 'lvf']
# One-hot encode categorical variables
for value in categorical_x_variables:
one_hot = pd.get_dummies(fix_chol[value], prefix=value)
fix_chol = fix_chol.join(one_hot)
fix_chol = fix_chol.drop(columns=value)
# Create list of x variables
x_variables = [x for x in list(fix_chol) if x != impute_variable]
# Create DataFrame with missing value(s) - will predict on
predict = fix_chol.loc[fix_chol[impute_variable]==-9]
# Set x and y predict DataFrames
predict_x, predict_y = predict[x_variables], predict[impute_variable]
# Create DataFrame to train on
train = fix_chol.loc[~(fix_chol[impute_variable]==-9)]
# Set x and y train DataFrames
train_x, train_y = train[x_variables], train[impute_variable]
# Fit scaler on train_x
train_x = scaler.fit_transform(train_x)
# Transform predict_x
predict_x = scaler.transform(predict_x)
# Obtain k (number of neighbors) by using sqrt(n)
k = round(sqrt(len(train_x)))
print(f"k is {k}.")
# Check to make sure k is odd number
if divmod(k, 2)[1] == 1:
print("k is an odd number. Good to proceed.")
else:
print("Need to make k an odd number.")
# Substract one to make k odd number
k -= 1
print(f"k is now {k}.")
# Predict value for predict_y
chol_prediction = KNeighborsRegressor(n_neighbors=k, metric='minkowski', weights='distance').fit(train_x, train_y).predict(predict_x)
print("The predictions for chol are:")
print(chol_prediction)
# Round chol_prediction to integer
for i in range(0, len(chol_prediction)):
chol_prediction[i] = round(number=chol_prediction[i])
print(f"The prediction for chol_prediction [{str(i)}] has been rounded to {chol_prediction[i]}.")
# Supply prediction back to appropriate patient
hungarian.loc[hungarian[impute_variable]==-9, impute_variable] = chol_prediction
Set Target Variable to Binary Range
# Set y variable to 0-1 range (as previous studies have done)
hungarian.loc[hungarian.num > 0, "num"] = 1
Exploratory Data Analysis
Determine Appropriate Alpha Value and Feature Engineering
# Determine 'strong' alpha value based on sample size
sample_size_one, strong_alpha_value_one = 100, 0.001
sample_size_two, strong_alpha_value_two = 1000, 0.0003
slope = (strong_alpha_value_two - strong_alpha_value_one)/(sample_size_two - sample_size_one)
strong_alpha_value = slope * (hungarian.shape[0] - sample_size_one) + strong_alpha_value_one
print(f"The alpha value for use in hypothesis tests is {strong_alpha_value}.")
# List of continuous variables
continuous_variables = ['age', 'trestbps', 'chol', 'thaldur', 'met', 'thalach', 'thalrest', 'tpeakbps', 'tpeakbpd',
'trestbpd', 'oldpeak', 'rldv5', 'rldv5e']
# List of categorical variables
categorical_variables = ['sex', 'painloc', 'painexer', 'relrest', 'cp', 'htn', 'fbs', 'restecg', 'prop', 'nitr',
'pro', 'diuretic', 'proto', 'exang', 'lvx3', 'lvx4', 'lvf']
# Target variable
target_variable = 'num'
### Feature engineering ###
# Create column of time between ekg and cardiac cath
# Create column of ekg dates
ekg_date = []
for year, month, day in zip(hungarian.ekgyr, hungarian.ekgmo, hungarian.ekgday):
x = str(year) + '-' + str(month) + '-' + str(day)
ekg_date.append(dt.datetime.strptime(x, '%y-%m-%d').strftime('%Y-%m-%d'))
# Append list to datetime to create column
hungarian['ekg_date'] = ekg_date
# Correct 2-30-86 issue (1986 was not a leap year)
hungarian.loc[(hungarian.cyr==86) & (hungarian.cmo==2) & (hungarian.cday==30), ('cmo', 'cday')] = (3,1)
cardiac_cath_date = []
for year, month, day in zip(hungarian.cyr, hungarian.cmo, hungarian.cday):
x = str(year) + '-' + str(month) + '-' + str(day)
print(x)
cardiac_cath_date.append(dt.datetime.strptime(x, '%y-%m-%d').strftime('%Y-%m-%d'))
# Append list to datetime to create column
hungarian['cardiac_cath_date'] = cardiac_cath_date
# Days between cardiac cath and ekg
hungarian['days_between_c_ekg'] = (pd.to_datetime(hungarian.cardiac_cath_date) - pd.to_datetime(hungarian.ekg_date)).dt.days
# Append days between cardiac cath and ekg to continuous variable list
continuous_variables.append('days_between_c_ekg')
# Create PCA variable from rldv5 and rldv5e
hungarian['rldv5_rldv5e_pca'] = PCA(n_components=1).fit_transform(hungarian[['rldv5', 'rldv5e']])
# Append new PCA'd variable to continuous variable list
continuous_variables.append('rldv5_rldv5e_pca')
# Dicitionary with continuous variable as key and spelled out version of variablea as value
continuous_variables_spelled_out_dict = {'age': 'Age', 'trestbps': 'Resting Blood Pressure (On Admission)',
'chol': 'Serum Cholesterol', 'thaldur': 'Duration of Exercise Test (Minutes)',
'met': 'METs Achieved', 'thalach': 'Maximum Heart Rate Achieved',
'thalrest': 'Resting Heart Rate',
'tpeakbps': 'Peak Exercise Blood Pressure (Systolic)',
'tpeakbpd': 'Peak Exercise Blood Pressure (Diastolic)',
'trestbpd': 'Resting Blood Pressure',
'oldpeak': 'ST Depression Induced by Exercise Relative to Rest',
'rldv5': 'Height at Rest',
'rldv5e': 'Height at Peak Exercise',
'days_between_c_ekg': 'Days Between Cardiac Catheterization and Electrocardiogram',
'rldv5_rldv5e_pca': "PCA variable for 'Height at Rest' and 'Height at Peak Exercise'"}
Heatmap of Continous Predictor Variables
# Heatmap of correlations
# Only return bottom portion of heatmap as top is duplicate and diagonal is redundant
continuous_variable_correlations = hungarian[continuous_variables].corr()
# Array of zeros with same shape as continuous_variable_correlations
mask = np.zeros_like(continuous_variable_correlations)
# Mark upper half and diagonal of mask as True
mask[np.triu_indices_from(mask)] = True
# Correlation heatmap
f, ax = plt.subplots()
f.subplots_adjust(left=0.31, right=1.06, top=0.96, bottom=0.43)
ax = sns.heatmap(hungarian[continuous_variables].corr(), cmap='PiYG', mask=mask, linewidths=.5, linecolor="white",
cbar=True)
ax.set_xticklabels(labels=continuous_variables_spelled_out_dict.values(),fontdict ={'fontweight': 'bold', 'fontsize':14},
rotation=45, ha="right",
rotation_mode="anchor")
ax.set_yticklabels(labels=continuous_variables_spelled_out_dict.values(),fontdict ={'fontweight': 'bold', 'fontsize':14})
ax.set_title("Heatmap of Continuous Predictor Features", fontdict ={'fontweight': 'bold', 'fontsize': 22})
DataFrame of Continuous Variable Correlations Greater Than 0.6 and Less Than -0.6 (More Numerical Alternative to Above Heatmap)
print(hungarian[continuous_variables].corr()[(hungarian[continuous_variables].corr()>0.6) | (hungarian[continuous_variables].corr()<-0.6)])
Histograms of Continuous Features by Target
fig, axes = plt.subplots(nrows=5, ncols=3)
fig.subplots_adjust(left=0.03, right=.97, top=0.90, bottom=0.03, hspace=0.7, wspace=0.25)
fig.suptitle('Distributions of Continuous Features by Target', fontweight='bold', fontsize= 22)
for ax, continuous in zip(axes.flatten(), continuous_variables):
for num_value in hungarian.num.unique():
ax.hist(hungarian.loc[hungarian.num == num_value, continuous], alpha=0.7, label=num_value)
ax.set_title(continuous_variables_spelled_out_dict[continuous], fontdict ={'fontweight': 'bold', 'fontsize': 15})
handles, legends = ax.get_legend_handles_labels()
legends_spelled_out_dict = {0: "No Presence of Heart Disease", 1: "Presence of Heart Disease"}
fig.legend(handles, legends_spelled_out_dict.values(), loc='upper left', bbox_to_anchor=(0.70, 1), prop={'weight': 'bold'})
Normality Tests
# Check normality of continuous variables
for continuous in continuous_variables:
print(continuous)
print(f"Kurtosis value: {stats.kurtosis(a=hungarian[continuous], fisher=True)}")
print(f"Sknewness value: {stats.skew(a=hungarian[continuous])}")
print(f"P-value from normal test: {stats.normaltest(a=hungarian[continuous])[1]}")
if stats.normaltest(a=hungarian[continuous])[1] < strong_alpha_value:
print("Reject null hypothesis the samples comes from a normal distribution.")
print("-------------------------------------------------------------------")
try:
print(f"Kurtosis value: {stats.kurtosis(a=stats.boxcox(x=hungarian[continuous])[0], fisher=True)}")
print(f"Sknewness value: {stats.skew(a=stats.boxcox(x=hungarian[continuous])[0])}")
print(f"P-value from normal test: {stats.normaltest(a=stats.boxcox(x=hungarian[continuous])[0])[1]}")
except ValueError as a:
if str(a) == "Data must be positive.":
print(f"{continuous} contains zero or negative values.")
else:
print("Do not reject the null hypothesis")
print('\n')
Data Transformations (Box-Cox Transformation)
# Boxcox necessary variables that reject the null hypothesis from normaltest in scipy.stats
hungarian['trestbps_boxcox'] = stats.boxcox(x=hungarian.trestbps)[0]
hungarian['chol_boxcox'] = stats.boxcox(x=hungarian.chol)[0]
hungarian['thalrest_boxcox'] = stats.boxcox(x=hungarian.thalrest)[0]
# Add boxcox'd variables to continuous_variables_spelled_out_dict
for boxcox_var in filter(lambda x: '_boxcox' in x, hungarian.columns):
continuous_variables_spelled_out_dict[boxcox_var] = continuous_variables_spelled_out_dict[
boxcox_var.split("_")[0]] + " Box-Cox"
Data Transformation of Serum Cholesterol - Distributions with KDE Overlaid
# Compare original distribution with boxcox'd distribution for chol
fig, axes = plt.subplots(nrows=1, ncols=2, sharey=True)
fig.subplots_adjust(left=0.05, right=.99, top=0.90, bottom=0.04)
fig.suptitle('Distributions with Kernel Density Estimation (KDE) Overlaid ', fontweight='bold', fontsize=26)
for ax, variable in zip(axes.flatten(), ['chol', 'chol_boxcox']):
print(ax, variable)
ax.hist(hungarian[variable])
ax2 = hungarian[variable].plot.kde(ax=ax, secondary_y=True, linewidth=8, alpha=0.8)
ax2.grid(False)
ax2.set_yticks([])
ax2.set_title(continuous_variables_spelled_out_dict[variable], fontdict={'fontweight': 'bold', 'fontsize': 24})
ax.text(0.78, 0.85, f"Kurtosis value: {'{:.3}'.format(stats.kurtosis(a=hungarian[variable], fisher=True))}\n"
f"Skewness value: {'{:.3}'.format(stats.skew(a=hungarian[variable]))}",
horizontalalignment='center', verticalalignment='center', transform=ax.transAxes,
bbox=dict(facecolor='none', edgecolor='black', pad=10.0, linewidth=4), weight='bold', fontsize=17)
ax.set_ylabel('Density', fontdict={'fontweight': 'bold', 'fontsize': 22})
# Expand figure to desired size first before running below code (this makes xtick labels appear and then can therefore be bolded)
for i in range(len(axes)):
axes[i].set_xticklabels(axes[i].get_xticklabels(), fontweight='bold', fontsize=18)
axes[0].set_yticklabels(axes[0].get_yticklabels(), fontweight='bold', fontsize=18)
Histograms of Continuous Features and Their Box-Cox’d Versions by Target
# Plot original and boxcox'd distributions to each other and against num
# Create list of boxcox'd variables and their originals
variables_for_inspection = list(itertools.chain.from_iterable([[x, x.split("_")[0]] for x in list(hungarian) if 'boxcox' in x]))
# Sort list in ascending order
variables_for_inspection.sort(reverse=False)
fig, axes = plt.subplots(nrows=len([x for x in variables_for_inspection if 'boxcox' in x]), ncols=2, figsize=(28,8))
fig.subplots_adjust(hspace=0.5)
fig.suptitle("Distributions of Continuous Features and their Box-Cox'd Versions", fontweight='bold', fontsize=20)
for ax, variable in zip(axes.flatten(), variables_for_inspection):
for num_value in hungarian.num.unique():
ax.hist(hungarian.loc[hungarian.num == num_value, variable], alpha=0.7, label=num_value)
ax.set_title(continuous_variables_spelled_out_dict[variable], fontdict={'fontweight': 'bold', 'fontsize': 24})
handles, legends = ax.get_legend_handles_labels()
legends_spelled_out_dict = {0: "No Presence of Heart Disease", 1: "Presence of Heart Disease"}
fig.legend(handles, legends_spelled_out_dict.values(), loc='upper left', bbox_to_anchor=(0.77, 1.0),
prop={'weight': 'bold', 'size': 14})
Chi-Square Tests
# Pearson chi-square tests
chi_square_analysis_list = []
for categorical in categorical_variables:
chi, p, dof, expected = stats.chi2_contingency(pd.crosstab(index=hungarian[categorical], columns=hungarian[target_variable]))
print(f"The chi-square value for {categorical} and {target_variable} is {chi}, and the p-value is" f" {p}, respectfully.")
chi_square_analysis_list.append([categorical, target_variable, chi, p])
# Create DataFrame from lists of lists
chi_square_analysis_df = pd.DataFrame(chi_square_analysis_list, columns=['variable', 'target', 'chi',
'p_value']).sort_values(by='p_value', ascending=True)
# Determine categorical variables that reject null
chi_square_analysis_df.loc[chi_square_analysis_df.p_value <= strong_alpha_value]
Contingency Tables, Odds Ratios, Descriptive Statistics, Data Visualizations, Chi-Square Tests
# Crosstab of age and num
pd.crosstab(index=hungarian.age,columns=hungarian.num, normalize=True)
# Distribution plot of age of all patients
plt.figure()
sns.distplot(hungarian['age'], kde=True, fit=stats.norm, rug=False,
kde_kws={"label": "Kernel Density Esimation (KDE)"},
fit_kws={"label": "Normal Distribution"}).set_title("Age Distribution of Patients")
plt.legend(loc='best')
plt.show()
# Statistical understanding of age of all patients
print(f"Mean +/- std of {hungarian['age'].name}: {round(hungarian['age'].describe()['mean'],2)} +/"
f" {round(hungarian['age'].describe()['std'],2)}. This means 68% of my patients lie between the ages of"
f" {round(hungarian['age'].describe()['mean'] - hungarian['age'].describe()['std'],2)} and"
f" {round(hungarian['age'].describe()['mean'] + hungarian['age'].describe()['std'],2)}.")
standard_devations = 2
print(f"Mean +/- {standard_devations} std of {hungarian['age'].name}: {round(hungarian['age'].describe()['mean'],2)} +/"
f" {round(hungarian['age'].describe()['std'] * standard_devations,2)}. This means 95% of my patients lie between the ages of"
f" {round(hungarian['age'].describe()['mean'] - (standard_devations * hungarian['age'].describe()['std']),2)} and"
f" {round(hungarian['age'].describe()['mean'] + (standard_devations * hungarian['age'].describe()['std']),2)}.")
standard_devations = 3
print(f"Mean +/- {standard_devations} std of {hungarian['age'].name}: {round(hungarian['age'].describe()['mean'],2)} +/"
f" {round(hungarian['age'].describe()['std'] * standard_devations,2)}. This means 99.7% of my patients lie between the ages of"
f" {round(hungarian['age'].describe()['mean'] - (standard_devations * hungarian['age'].describe()['std']),2)} and"
f" {round(hungarian['age'].describe()['mean'] + (standard_devations * hungarian['age'].describe()['std']),2)}.")
print(f"Mode of {hungarian['age'].name}: {hungarian['age'].mode()[0]}\nMedian of {hungarian['age'].name}: {hungarian['age'].median()}")
# Distribution plot of age of patients broken down by sex - female
plt.figure()
sns.distplot(hungarian.loc[hungarian['sex']==0, 'age'], kde=True, fit=stats.norm,
kde_kws={"label": "Kernel Density Esimation (KDE)"},
fit_kws={"label": "Normal Distribution"}).set_title('Female Age Distribution')
plt.legend(loc='best')
plt.show()
# Women age information
print(f"Mean +/- std of {hungarian.loc[hungarian['sex']==0, 'age'].name} for women: "
f"{round(hungarian.loc[hungarian['sex']==0, 'age'].describe()['mean'],2)} +/ "
f"{round(hungarian.loc[hungarian['sex']==0, 'age'].describe()['std'],2)}")
print(f"Mode of {hungarian.loc[hungarian['sex']==0, 'age'].name} for women: "
f"{hungarian.loc[hungarian['sex']==0, 'age'].mode()[0]}\nMedian of "
f"{hungarian.loc[hungarian['sex']==0, 'age'].name} for women: + "
f"{hungarian.loc[hungarian['sex']==0, 'age'].median()}")
print('\n')
# Distribution plot of age of patients broken down by sex - male
plt.figure()
sns.distplot(hungarian.loc[hungarian['sex']==1, 'age'],kde=True, fit=stats.norm,
kde_kws={"label": "Kernel Density Esimation (KDE)"},
fit_kws={"label": "Normal Distribution"}).set_title('Male Age Distribution')
plt.legend(loc='best')
plt.show()
# Men age information
print(f"Mean +/- std of {hungarian.loc[hungarian['sex']==1, 'age'].name} for men: "
f"{round(hungarian.loc[hungarian['sex']==1, 'age'].describe()['mean'],2)} +/ "
f"{round(hungarian.loc[hungarian['sex']==1, 'age'].describe()['std'],2)}")
print(f"Mode of {hungarian.loc[hungarian['sex']==1, 'age'].name} for men: "
f"{hungarian.loc[hungarian['sex']==1, 'age'].mode()[0]}\nMedian of "
f"{hungarian.loc[hungarian['sex']==1, 'age'].name} for men: + "
f"{hungarian.loc[hungarian['sex']==1, 'age'].median()}")
## Sex
# Get counts of sex
print(hungarian.sex.value_counts())
print(f'The hungarian dataset consists of {hungarian.sex.value_counts()[0]} females and'
f' {hungarian.sex.value_counts()[1]} males.')
# Bar graph of sex by num
plt.figure()
sex_dict = {0: "female", 1: "male"}
sns.countplot(x="sex", hue="num", data=hungarian).set(title='Heart Disease Indicator by Sex', xticklabels=sex_dict.values())
plt.show()
# Crosstab of sex by num
print(pd.crosstab(index=hungarian.sex, columns=hungarian.num))
# Crosstab of sex by num - all values normalized
# Of all patients in dataset, 32% were males that had heart disease. 4% were females that had heart disease.
print(pd.crosstab(index=hungarian.sex, columns=hungarian.num, normalize='all'))
# Crosstab of sex by num - rows normalized
# 15% of females had heart disease. 44% of males had heart disease.
print(pd.crosstab(index=hungarian.sex, columns=hungarian.num, normalize='index'))
# Crosstab of sex by num - columns normalized
# 89% of the patients with heart disease were males. 11% were females.
print(pd.crosstab(index=hungarian.sex, columns=hungarian.num, normalize='columns'))
# Contingency table of sex by num
contingency = pd.crosstab(index=hungarian.sex, columns=hungarian.num)
print(contingency)
# Pearson chi-square test
chi, p, dof, expected = stats.chi2_contingency(contingency)
if p <= strong_alpha_value:
print(f"Reject the null hypothesis of no association between {contingency.index.name} and diagnosis of heart "
f"disease and conclude there is an association between {contingency.index.name} and diagnosis of heart "
f"disease. The probability of a heart disease diagnosis is not the same for male and female patients.")
else:
print(f"Fail to reject the null of no association between sex and diagnosis of heart disease. The probability of a "
f"heart disease diagnosis is the same regardless of a patient's sex.")
# Compute odds ratio and risk ratio
table = sm.stats.Table2x2(contingency)
print(table.summary())
print(f"The odds ratio is {table.oddsratio}. This means males are {round(table.oddsratio,2)} times more likely to be "
f"diagnosed with heart disease than females.")
## Painloc
# Bar graph of painloc by num
plt.figure()
painloc_dict = {0: "otherwise", 1: "substernal"}
sns.countplot(x="painloc", hue="num", data=hungarian).set(title='Heart Disease Indicator by Pain Location', xticklabels=painloc_dict.values())
plt.show()
# Contingency table of painloc by num
contingency = pd.crosstab(index=hungarian.painloc, columns=hungarian.num)
print(contingency)
# Pearson chi-square test
chi, p, dof, expected = stats.chi2_contingency(contingency)
print(f"The chi-square value for {contingency.index.name} and {contingency.columns.name} is {chi}, and the p-value is"
f" {p}, respectfully.")
if p <= strong_alpha_value:
print(f"Reject the null hypothesis of no association between {contingency.index.name} and diagnosis of heart "
f"disease and conclude there is an association between {contingency.index.name} and diagnosis of heart "
f"disease. The probability of a heart disease diagnosis is not the same based on chest pain location.")
else:
print(f"Fail to reject the null of no association between {contingency.index.name} and diagnosis of heart disease. "
f"The probability of a heart disease diagnosis is the same regardless of chest pain location.")
# Compute odds ratio and risk ratio
table = sm.stats.Table2x2(contingency)
print(table.summary())
print(f"The odds ratio is {table.oddsratio}.")
## Painexer
# Bar graph of painexer by num
plt.figure()
painexer_dict = {0: "otherwise", 1: "provoked by exertion"}
sns.countplot(x="painexer", hue="num",
data=hungarian).set(title='Heart Disease Indicator by Pain Exertion', xticklabels=painexer_dict.values())
plt.show()
# Contingency table of painexer by num
contingency = pd.crosstab(index=hungarian.painexer, columns=hungarian.num)
print(contingency)
# Pearson chi-square test
chi, p, dof, expected = stats.chi2_contingency(contingency)
print(f"The chi-square value for {contingency.index.name} and {contingency.columns.name} is {chi}, and the p-value is"
f" {p}, respectfully. The expected values are\n{expected}.")
if p <= strong_alpha_value:
print(f"Reject the null hypothesis of no association between {contingency.index.name} and diagnosis of heart "
f"disease and conclude there is an association between {contingency.index.name} and diagnosis of heart "
f"disease. The probability of a heart disease diagnosis is not the same based on how chest pain is provoked.")
else:
print(f"Fail to reject the null of no association between {contingency.index.name} and diagnosis of heart disease. "
f"The probability of a heart disease diagnosis is the same regardless of how chest pain is provoked.")
# Compute odds ratio and risk ratio
table = sm.stats.Table2x2(contingency)
print(table.summary())
print(f"The odds ratio is {table.oddsratio}. This means patients with their chest pain provoked by exertion are "
f"{round(table.oddsratio,2)} times more likely to have a diagnosis of heart disease than those patients with "
f"their chest pain provoked otherwise.")
## Relrest
# Bar graph of relrest by num
plt.figure()
relrest_dict = {0: "otherwise", 1: "relieved after rest"}
sns.countplot(x="relrest", hue="num", data=hungarian).set(title='Heart Disease Indicator by Pain Relief', xticklabels=relrest_dict.values())
plt.show()
# Contingency table of relrest by num
contingency = pd.crosstab(index=hungarian.relrest, columns=hungarian.num)
print(contingency)
# Pearson chi-square test
chi, p, dof, expected = stats.chi2_contingency(contingency)
print(f"The chi-square value for {contingency.index.name} and {contingency.columns.name} is {chi}, and the p-value is"
f" {p}, respectfully. The expected values are\n{expected}.")
if p <= strong_alpha_value:
print(f"Reject the null hypothesis of no association between {contingency.index.name} and diagnosis of heart "
f"disease and conclude there is an association between {contingency.index.name} and diagnosis of heart "
f"disease. The probability of a heart disease diagnosis is not the same for pain relieved after rest and "
f"otherwise.")
else:
print(f"Fail to reject the null of no association between {contingency.index.name} and diagnosis of heart disease. "
f"The probability of a heart disease diagnosis is the same regardless of when the pain is relieved.")
# Compute odds ratio and risk ratio
table = sm.stats.Table2x2(contingency)
print(table.summary())
print(f"The odds ratio is {table.oddsratio}. This means patients with their chest pain relieved after rest are "
f"{round(table.oddsratio,2)} times more likely to have a diagnosis of heart disease than those patients with "
f"their chest pain relieved otherwise.")
## Cp
# Bar graph of cp by num
plt.figure()
cp_dict = {1: "typical angina", 2: "atypical angina", 3: "non-anginal pain", 4: "asymptomatic"}
sns.countplot(x="cp", hue="num", data=hungarian).set(title='Heart Disease Indicator by Chest Pain Type', xticklabels=cp_dict.values())
plt.show()
# Contingency table of cp by cum
contingency = pd.crosstab(index=hungarian.cp, columns=hungarian.num)
print(contingency)
# Pearson chi-square test
chi, p, dof, expected = stats.chi2_contingency(contingency)
print(f"The chi-square value for {contingency.index.name} and {contingency.columns.name} is {chi}, and the p-value is"
f" {p}, respectfully.")
if p <= strong_alpha_value:
print(f"Reject the null hypothesis of no association between {contingency.index.name} and diagnosis of heart "
f"disease and conclude there is an association between {contingency.index.name} and diagnosis of heart "
f"disease. The probability of a heart disease diagnosis is not the same depending on chest pain type.")
else:
print(
f"Fail to reject the null of no association between {contingency.index.name} and diagnosis of heart disease. "
f"The probability of a heart disease diagnosis is the same regardless of chest pain type.")
Feature Engineering
hungarian["thalach_div_by_thalrest"] = hungarian["thalach"]/hungarian["thalrest"]
hungarian["tpeakbps_div_by_tpeakbpd"] = hungarian["tpeakbps"]/hungarian["tpeakbpd"]
hungarian["thaldur_div_by_met"] = hungarian["thaldur"]/hungarian["met"]
hungarian["chol_div_by_age"] = hungarian["chol"]/hungarian["age"]
hungarian["chol_div_by_met"] = hungarian["chol"]/hungarian["met"]
hungarian["chol_div_by_thalach"] = hungarian["chol"]/hungarian["thalach"]
hungarian["chol_div_by_thalrest"] = hungarian["chol"]/hungarian["thalrest"]
hungarian["thalrest_div_by_rldv5"] = hungarian["thalrest"]/hungarian["rldv5"]
hungarian["thalach_div_by_rldv5e"] = hungarian["thalrest"]/hungarian["rldv5e"]
hungarian["trestbps_boxcox_div_by_tpeakbpd"] = hungarian["trestbps_boxcox"]/hungarian["tpeakbpd"]
hungarian["chol_boxcox_div_by_age"] = hungarian["chol_boxcox"]/hungarian["age"]
hungarian["chol_boxcox_div_by_met"] = hungarian["chol_boxcox"]/hungarian["met"]
hungarian["chol_boxcox_div_by_thalach"] = hungarian["chol_boxcox"]/hungarian["thalach"]
hungarian["chol_boxcox_div_by_thalrest"] = hungarian["chol_boxcox"]/hungarian["thalrest"]
hungarian["thalach_div_by_thalrest_boxcox"] = hungarian["thalach"]/hungarian["thalrest_boxcox"]
hungarian["chol_div_by_thalrest_boxcox"] = hungarian["chol"]/hungarian["thalrest_boxcox"]
hungarian["thalrest_boxcox_div_by_rldv5"] = hungarian["thalrest_boxcox"]/hungarian["rldv5"]
# Bin age
hungarian['agebinned'] = pd.cut(x=hungarian.age, bins=5, labels = ['0', '1', '2', '3', '4'])
DataFrame of Continuous Variable Correlations Greater Than 0.6 and Less Than 1.0 and Correlations Less Than -0.6 and Greater Than -1.0 With Entirely Null Columns Dropped - All Feature Engineered Variables Included
# Add boxcox'd variables to continuous_variables list
continuous_variables.extend([x for x in list(hungarian) if 'boxcox' in x])
# Add interaction variables to continuous_variables list
continuous_variables.extend([x for x in list(hungarian) if 'div_by' in x])
# Correlations > 0.6 and < 1.0 and <-0.6 and >-1.0, drop all null columns
hungarian[continuous_variables].corr()[((hungarian[continuous_variables].corr() > 0.6) &
(hungarian[continuous_variables].corr() < 1.0)) |
((hungarian[continuous_variables].corr()<-0.6) &
(hungarian[continuous_variables].corr()>-1.0))].dropna(axis=1, how='all')
Go Back to Exploratory Data Analysis
Model Building
DataFrame to Append Model Results
# Create empty DataFrame to append all model results to
all_model_results = pd.DataFrame()
# Create DataFrame to append top model results to
top_model_results = pd.DataFrame(columns=['model_type', 'solver', 'best_model_params_grid_search', 'best_score_grid_search',
'true_negatives', 'false_positives', 'false_negatives', 'true_positives',
'recall', 'precision', 'f1_score', 'variables_not_used', 'variables_used',
'model_params_grid_search'])
Copy of hungarian DataFrame for Modeling and Save Target Variable for Later Use
model = hungarian.copy()
# Save target variable for use with pickle files
model['num'].to_pickle("hungarian_target_variable.pkl")
Create Unique Set of Variables
# Drop columns
variables_to_drop_for_modeling_one = ['id', 'ekgyr', 'ekgmo', 'ekgday', 'cyr', 'cmo', 'cday', 'lvx3', 'lvx4', 'lvf',
'proto', 'ekg_date', 'cardiac_cath_date', 'rldv5_rldv5e_pca',
'days_between_c_ekg', 'trestbps_boxcox', 'chol_boxcox', 'thalrest_boxcox',
'thalach_div_by_thalrest', 'tpeakbps_div_by_tpeakbpd', 'thaldur_div_by_met',
'chol_div_by_met', 'chol_div_by_thalach', 'chol_div_by_thalrest',
'chol_div_by_age', 'thalrest_div_by_rldv5', 'thalach_div_by_rldv5e', 'agebinned',
'trestbps_boxcox_div_by_tpeakbpd', 'chol_boxcox_div_by_age',
'chol_boxcox_div_by_met', 'chol_boxcox_div_by_thalach',
'chol_boxcox_div_by_thalrest', 'thalach_div_by_thalrest_boxcox',
'chol_div_by_thalrest_boxcox', 'thalrest_boxcox_div_by_rldv5']
# Define categorical variables
categorical_variables_for_modeling_one = ['sex', 'painloc', 'painexer', 'relrest', 'cp', 'htn', 'fbs', 'restecg',
'prop', 'nitr', 'pro', 'diuretic', 'exang']
# Drop columns
variables_to_drop_for_modeling_two = ['id', 'chol', 'thalrest', 'trestbps', 'ekgyr', 'ekgmo', 'ekgday', 'cyr', 'cmo',
'cday', 'ekg_date', 'cardiac_cath_date', 'rldv5', 'lvx3', 'lvx4', 'lvf', 'pro',
'proto', 'rldv5_rldv5e_pca', 'thalach_div_by_thalrest',
'tpeakbps_div_by_tpeakbpd', 'thaldur_div_by_met', 'chol_div_by_met',
'chol_div_by_thalach', 'chol_div_by_thalrest', 'chol_div_by_age',
'thalrest_div_by_rldv5', 'thalach_div_by_rldv5e', 'agebinned',
'trestbps_boxcox_div_by_tpeakbpd', 'chol_boxcox_div_by_age',
'chol_boxcox_div_by_met', 'chol_boxcox_div_by_thalach',
'chol_boxcox_div_by_thalrest', 'thalach_div_by_thalrest_boxcox',
'chol_div_by_thalrest_boxcox', 'thalrest_boxcox_div_by_rldv5']
# Define categorical variables
categorical_variables_for_modeling_two = ['sex', 'painloc', 'painexer', 'relrest', 'cp', 'htn', 'fbs', 'restecg',
'prop', 'nitr', 'diuretic', 'exang']
# Drop columns
variables_to_drop_for_modeling_three = ['id', 'chol', 'thalrest', 'trestbps', 'ekgyr', 'ekgmo', 'ekgday', 'cyr', 'cmo',
'cday', 'ekg_date', 'cardiac_cath_date', 'lvx3', 'lvx4', 'lvf', 'proto',
'rldv5_rldv5e_pca', 'thalach_div_by_thalrest',
'tpeakbps_div_by_tpeakbpd', 'thaldur_div_by_met', 'chol_div_by_met',
'chol_div_by_thalach', 'chol_div_by_thalrest', 'chol_div_by_age',
'thalrest_div_by_rldv5', 'thalach_div_by_rldv5e', 'agebinned',
'trestbps_boxcox_div_by_tpeakbpd', 'chol_boxcox_div_by_age',
'chol_boxcox_div_by_met', 'chol_boxcox_div_by_thalach',
'chol_boxcox_div_by_thalrest', 'thalach_div_by_thalrest_boxcox',
'chol_div_by_thalrest_boxcox', 'thalrest_boxcox_div_by_rldv5']
# Define categorical variables
categorical_variables_for_modeling_three = ['sex', 'painloc', 'painexer', 'relrest', 'cp', 'htn', 'fbs', 'restecg', 'prop',
'nitr', 'diuretic', 'exang', 'pro']
# Drop columns
variables_to_drop_for_modeling_four = ['id', 'ekgyr', 'ekgmo', 'ekgday', 'cyr', 'cmo', 'cday', 'lvx3', 'lvx4', 'lvf',
'proto', 'ekg_date', 'cardiac_cath_date', 'rldv5', 'rldv5e', 'trestbps_boxcox',
'chol_boxcox', 'thalrest_boxcox', 'agebinned',
'trestbps_boxcox_div_by_tpeakbpd', 'chol_boxcox_div_by_age',
'chol_boxcox_div_by_met', 'chol_boxcox_div_by_thalach',
'chol_boxcox_div_by_thalrest', 'thalach_div_by_thalrest_boxcox',
'chol_div_by_thalrest_boxcox', 'thalrest_boxcox_div_by_rldv5']
# Define categorical variables
categorical_variables_for_modeling_four = ['sex', 'painloc', 'painexer', 'relrest', 'cp', 'htn', 'fbs', 'restecg', 'prop',
'nitr', 'diuretic', 'exang', 'pro']
# Drop columns
variables_to_drop_for_modeling_five = ['id', 'age', 'ekgyr', 'ekgmo', 'ekgday', 'cyr', 'cmo', 'cday', 'lvx3', 'lvx4', 'lvf',
'proto', 'ekg_date', 'cardiac_cath_date','rldv5_rldv5e_pca', 'trestbps_boxcox',
'chol_boxcox', 'thalrest_boxcox', 'trestbps_boxcox_div_by_tpeakbpd',
'chol_boxcox_div_by_age', 'chol_boxcox_div_by_met', 'chol_boxcox_div_by_thalach',
'chol_boxcox_div_by_thalrest', 'thalach_div_by_thalrest_boxcox',
'chol_div_by_thalrest_boxcox', 'thalrest_boxcox_div_by_rldv5']
# Define categorical variables
categorical_variables_for_modeling_five = ['sex', 'painloc', 'painexer', 'relrest', 'cp', 'htn', 'fbs', 'restecg', 'prop',
'nitr', 'diuretic', 'exang', 'pro', 'agebinned']
# Drop columns
variables_to_drop_for_modeling_six = ['id', 'age', 'chol', 'thalrest', 'trestbps', 'ekgyr', 'ekgmo', 'ekgday', 'cyr',
'cmo', 'cday', 'lvx3', 'lvx4', 'lvf',
'proto', 'ekg_date', 'cardiac_cath_date','rldv5_rldv5e_pca',
'thalach_div_by_thalrest', 'tpeakbps_div_by_tpeakbpd', 'thaldur_div_by_met',
'chol_div_by_met', 'chol_div_by_thalach', 'chol_div_by_thalrest',
'chol_div_by_age', 'thalrest_div_by_rldv5', 'thalach_div_by_rldv5e']
# Define categorical variables
categorical_variables_for_modeling_six = ['sex', 'painloc', 'painexer', 'relrest', 'cp', 'htn', 'fbs', 'restecg', 'prop',
'nitr', 'diuretic', 'exang', 'pro', 'agebinned']
# Drop columns
variables_to_drop_for_modeling_seven = ['id', 'chol', 'thalrest', 'trestbps', 'ekgyr', 'ekgmo', 'ekgday', 'cyr',
'cmo', 'cday', 'lvx3', 'lvx4', 'lvf',
'proto', 'ekg_date', 'cardiac_cath_date','rldv5_rldv5e_pca',
'thalach_div_by_thalrest', 'tpeakbps_div_by_tpeakbpd', 'thaldur_div_by_met',
'chol_div_by_met', 'chol_div_by_thalach', 'chol_div_by_thalrest',
'chol_div_by_age', 'thalrest_div_by_rldv5', 'thalach_div_by_rldv5e', 'agebinned']
# Define categorical variables
categorical_variables_for_modeling_seven = ['sex', 'painloc', 'painexer', 'relrest', 'cp', 'htn', 'fbs', 'restecg', 'prop',
'nitr', 'diuretic', 'exang', 'pro']
# Make list of lists for variables to drop
variables_to_drop_list = [variables_to_drop_for_modeling_one, variables_to_drop_for_modeling_two,
variables_to_drop_for_modeling_three, variables_to_drop_for_modeling_four,
variables_to_drop_for_modeling_five,variables_to_drop_for_modeling_six,
variables_to_drop_for_modeling_seven]
# Make list of lists for categorical variables to model
categorical_variables_for_modeling_list = [categorical_variables_for_modeling_one,
categorical_variables_for_modeling_two,
categorical_variables_for_modeling_three,
categorical_variables_for_modeling_four,
categorical_variables_for_modeling_five,
categorical_variables_for_modeling_six,
categorical_variables_for_modeling_seven]
Logistic Regression
# Unique variable combination runs
for index, (vars_to_drop, cat_vars_to_model) in enumerate(zip(variables_to_drop_list,
categorical_variables_for_modeling_list), start=1):
print(f"Model run: {index}")
# Create copy of hungarian for regression modeling
model = hungarian.copy()
# Drop variables
model = model.drop(columns=vars_to_drop)
# Dummy variable categorical variables
model = pd.get_dummies(data=model, columns=cat_vars_to_model, drop_first=True)
# Create target variable
y = model['num']
# Create feature variables
x = model.drop(columns='num')
# Obtain recursive feature elimination values for all solvers and get average
# (not sure what to do about ConvergenceWarning - get warning but also get result for each solver)
rfe_logit = pd.DataFrame(data=list(x), columns=['variable'])
for solve in ['liblinear', 'newton-cg', 'lbfgs', 'sag', 'saga']:
rfe_logit = rfe_logit.merge(pd.DataFrame(data=[list(x), RFE(LogisticRegression(solver=solve, max_iter=100),
n_features_to_select=1).fit(x, y).ranking_.tolist()]).T.rename(columns={0: 'variable', 1:
'rfe_ranking_' + solve}), on='variable')
# Get average ranking for each variable
rfe_logit['rfe_ranking_avg'] = rfe_logit[['rfe_ranking_liblinear', 'rfe_ranking_newton-cg', 'rfe_ranking_lbfgs',
'rfe_ranking_sag', 'rfe_ranking_saga']].mean(axis=1)
# Sort DataFrame
rfe_logit = rfe_logit.sort_values(by='rfe_ranking_avg', ascending=True).reset_index(drop=True)
# Train/test split
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.33, random_state=43)
# Run models - start at top and add variables with each iteration
# Test 'weaker' alpha value
strong_alpha_value = 0.04
model_search_logit = []
logit_variable_list = []
insignificant_variables_list = []
for i in range(len(rfe_logit)):
if rfe_logit['variable'][i] not in logit_variable_list and rfe_logit['variable'][i] not in insignificant_variables_list:
logit_variable_list.extend([rfe_logit['variable'][i]])
# logit_variable_list = list(set(logit_variable_list).difference(set(insignificant_variables_list)))
logit_variable_list = [x for x in logit_variable_list if x not in insignificant_variables_list]
print(logit_variable_list)
# Add related one-hot encoded variables if variable is categorical
if logit_variable_list[-1].split('_')[-1] in sorted([x for x in list(set([x.split('_')[-1] for x in list(x)])) if len(x) == 1]):
logit_variable_list.extend([var for var in list(x) if logit_variable_list[-1].split('_')[0] in var and var != logit_variable_list[-1]])
print(logit_variable_list)
# Build logistic regression
sm_logistic = sm.Logit(y_train, x_train[logit_variable_list]).fit()
# All p-values are significant
if all(p_values < strong_alpha_value for p_values in sm_logistic.summary2().tables[1]._getitem_column("P>|z|").values):
print("-*"*60)
print((sm_logistic.summary2().tables[1]._getitem_column("P>|z|").index.tolist(),
sm_logistic.summary2().tables[1]._getitem_column("P>|z|").values.tolist()))
print("-*"*60)
print("-*"*60)
model_search_logit.append([(sm_logistic.summary2().tables[0][0][6], sm_logistic.summary2().tables[0][1][6]),
(sm_logistic.summary2().tables[0][2][0], sm_logistic.summary2().tables[0][3][0]),
(sm_logistic.summary2().tables[0][2][1], sm_logistic.summary2().tables[0][3][1]),
(sm_logistic.summary2().tables[0][2][2], sm_logistic.summary2().tables[0][3][2]),
(sm_logistic.summary2().tables[0][2][3], sm_logistic.summary2().tables[0][3][3]),
(sm_logistic.summary2().tables[0][2][4], sm_logistic.summary2().tables[0][3][4]),
(sm_logistic.summary2().tables[0][2][5], sm_logistic.summary2().tables[0][3][5]),
(sm_logistic.summary2().tables[1]._getitem_column("P>|z|").index.tolist(),
sm_logistic.summary2().tables[1]._getitem_column("P>|z|").values.tolist())])
# P-value(s) of particular variable(s) is not significant
elif any(p_values > strong_alpha_value for p_values in sm_logistic.summary2().tables[1]._getitem_column("P>|z|").values):
print('*'*60)
print(logit_variable_list[-1])
print('*'*60)
if logit_variable_list[-1].split('_')[-1] in sorted([x for x in list(set([x.split('_')[-1] for x in list(x)])) if len(x) == 1]):
cat_var_level_check = sm_logistic.summary2().tables[1]._getitem_column("P>|z|")[sm_logistic.summary2().
tables[1]._getitem_column("P>|z|").index.isin([var for var in list(x) if
logit_variable_list[-1].split('_')[0] in var])]
# If True, at least one level of the categorical variable is significant so keep all levels of variable
if any(p_values < strong_alpha_value for p_values in cat_var_level_check.values):
model_search_logit.append([(sm_logistic.summary2().tables[0][0][6], sm_logistic.summary2().tables[0][1][6]),
(sm_logistic.summary2().tables[0][2][0], sm_logistic.summary2().tables[0][3][0]),
(sm_logistic.summary2().tables[0][2][1], sm_logistic.summary2().tables[0][3][1]),
(sm_logistic.summary2().tables[0][2][2], sm_logistic.summary2().tables[0][3][2]),
(sm_logistic.summary2().tables[0][2][3], sm_logistic.summary2().tables[0][3][3]),
(sm_logistic.summary2().tables[0][2][4], sm_logistic.summary2().tables[0][3][4]),
(sm_logistic.summary2().tables[0][2][5], sm_logistic.summary2().tables[0][3][5]),
(sm_logistic.summary2().tables[1]._getitem_column("P>|z|").index.tolist(),
sm_logistic.summary2().tables[1]._getitem_column("P>|z|").values.tolist())])
# Else False - remove all levels of categorical variable
else:
print("-"*60)
print(sm_logistic.summary2())
insignificant_variables_list.extend(cat_var_level_check.index)
else:
print('='*60)
print(sm_logistic.summary2())
print(logit_variable_list[-1])
cont_var_check = sm_logistic.summary2().tables[1]._getitem_column("P>|z|")[sm_logistic.summary2().
tables[1]._getitem_column("P>|z|").index.isin([logit_variable_list[-1]])]
# Continuous variable is significant
if cont_var_check.values[0] < strong_alpha_value:
model_search_logit.append([(sm_logistic.summary2().tables[0][0][6], sm_logistic.summary2().tables[0][1][6]),
(sm_logistic.summary2().tables[0][2][0], sm_logistic.summary2().tables[0][3][0]),
(sm_logistic.summary2().tables[0][2][1], sm_logistic.summary2().tables[0][3][1]),
(sm_logistic.summary2().tables[0][2][2], sm_logistic.summary2().tables[0][3][2]),
(sm_logistic.summary2().tables[0][2][3], sm_logistic.summary2().tables[0][3][3]),
(sm_logistic.summary2().tables[0][2][4], sm_logistic.summary2().tables[0][3][4]),
(sm_logistic.summary2().tables[0][2][5], sm_logistic.summary2().tables[0][3][5]),
(sm_logistic.summary2().tables[1]._getitem_column("P>|z|").index.tolist(),
sm_logistic.summary2().tables[1]._getitem_column("P>|z|").values.tolist())])
else:
print('^'*60)
print(logit_variable_list[-1])
insignificant_variables_list.append(logit_variable_list[-1])
# Create DataFrame of logisitic regression results
model_search_logit = pd.DataFrame(model_search_logit, columns = ['converged', 'pseudo_r_squared', 'aic', 'bic',
'log_likelihood', 'll_null', 'llr_p_value', 'columns_significance'])
model_results_logit = []
for solve in ['liblinear', 'newton-cg', 'lbfgs', 'sag', 'saga']:
for col in model_search_logit['columns_significance']:
print(solve, col[0])
try:
logit_predict = cross_val_predict(LogisticRegression(solver=solve, max_iter=100), x[col[0]], y, cv=5)
print(confusion_matrix(y_true=y, y_pred=logit_predict))
conf_matr = confusion_matrix(y_true=y, y_pred=logit_predict)
model_results_logit.append([solve, col[0], conf_matr[0][0], conf_matr[0][1], conf_matr[1][0], conf_matr[1][1]])
except ConvergenceWarning:
print("#"*60)
# Create DataFrame of results
model_results_logit = pd.DataFrame(model_results_logit, columns = ['solver', 'variables_used', 'true_negatives', 'false_positives',
'false_negatives', 'true_positives'])
# Create recall, precision, and f1-score columns
model_results_logit['recall'] = model_results_logit.true_positives/(model_results_logit.true_positives + model_results_logit.false_negatives)
model_results_logit['precision'] = model_results_logit.true_positives/(model_results_logit.true_positives + model_results_logit.false_positives)
model_results_logit['f1_score'] = 2 * (model_results_logit.precision * model_results_logit.recall) / (model_results_logit.precision + model_results_logit.recall)
# Sort DataFrame
model_results_logit = model_results_logit.sort_values(by=['f1_score'], ascending=False)
print(model_results_logit)
if len(model_results_logit.loc[model_results_logit.f1_score==model_results_logit.f1_score.max()]) > 1:
top_model_result_logit = model_results_logit.loc[(model_results_logit.f1_score == model_results_logit.f1_score.max()) &
(model_results_logit['variables_used'].apply(len) == min(map(lambda x: len(x[[1]][0]),
model_results_logit.loc[model_results_logit.f1_score==model_results_logit.f1_score.max()].values)))].sample(n=1)
else:
top_model_result_logit = model_results_logit.loc[model_results_logit.f1_score == model_results_logit.f1_score.max()]
top_model_results = top_model_results.append(other= top_model_result_logit, sort=False)
print(f"Top logit model: \n {top_model_result_logit}")
# Append top_model_result_logit results to all_model_results DataFrame
logit_predict_proba = cross_val_predict(LogisticRegression(solver=top_model_result_logit["solver"].values[0],
max_iter=100), x[top_model_result_logit["variables_used"].values[0]], y, cv=5, method="predict_proba")
all_model_results['logit_'+inflect.engine().number_to_words(index)+'_pred_zero'] = logit_predict_proba[:,0]
all_model_results['logit_'+inflect.engine().number_to_words(index)+'_pred_one'] = logit_predict_proba[:,1]
# Fill in model_type columns
top_model_results['model_type'] = top_model_results['model_type'].fillna(value='logit')
Random Forest
# Unique variable combination runs
for index, (vars_to_drop, cat_vars_to_model) in enumerate(zip(variables_to_drop_list,
categorical_variables_for_modeling_list), start=1):
print(f"Model run: {index}")
# Create copy of hungarian for non-regression modeling
model = hungarian.copy()
# Drop columns
model = model.drop(columns=vars_to_drop)
# Dummy variable categorical variables
model = pd.get_dummies(data=model, columns=cat_vars_to_model, drop_first=False)
# Create target variable
y = model['num']
# Create feature variables
x = model.drop(columns='num')
# Define parameters of Random Forest Classifier
random_forest_model = RandomForestClassifier(random_state=1)
# Define parameters for grid search
param_grid = {'n_estimators': np.arange(10, 111, step=5), 'criterion': ['gini', 'entropy'],
'max_features': np.arange(2, 25, step=3)}
cv = ShuffleSplit(n_splits=5, test_size=0.3)
# Define grid search CV parameters
grid_search = GridSearchCV(random_forest_model, param_grid, cv=cv) # , scoring='recall' # warm_start=True
# Loop to iterate through least important variables according to random_forest_feature_importance and grid search
x_all = list(x)
model_search_rfc = []
while True:
print("--------------------------------")
print(len(list(x)))
print(print(param_grid['max_features']))
print("--------------------------------")
# try:
grid_search.fit(x, y)
print(f'Best parameters for current grid seach: {grid_search.best_params_}')
print(f'Best score for current grid seach: {grid_search.best_score_}')
# Define parameters of Random Forest Classifier from grid search
random_forest_model = RandomForestClassifier(criterion=grid_search.best_params_['criterion'],
max_features=grid_search.best_params_['max_features'],
n_estimators=grid_search.best_params_['n_estimators'],
random_state=1)
# Cross-validate and predict using Random Forest Classifier
random_forest_predict = cross_val_predict(random_forest_model, x, y, cv=5)
print(confusion_matrix(y_true=y, y_pred=random_forest_predict))
conf_matr = confusion_matrix(y_true=y, y_pred=random_forest_predict)
model_search_rfc.append([grid_search.best_params_, grid_search.best_score_, conf_matr[0][0], conf_matr[0][1],
conf_matr[1][0], conf_matr[1][1], set(x_all).difference(x)])
# Run random forest with parameters from grid search to obtain feature importances
random_forest_feature_importance = pd.DataFrame(data=[list(x),
RandomForestClassifier(criterion=grid_search.best_params_['criterion'],
max_features=grid_search.best_params_['max_features'],
n_estimators=grid_search.best_params_['n_estimators'], random_state=1).fit(x,y).feature_importances_.tolist()]).T.rename(columns={0:'variable',
1:'importance'}).sort_values(by='importance', ascending=False)
print(random_forest_feature_importance)
if len(random_forest_feature_importance.loc[random_forest_feature_importance.importance<0.01]) > 0:
for i in range(1, len(random_forest_feature_importance.loc[random_forest_feature_importance.importance<0.01])+1):
print(f"'Worst' variable being examined: {random_forest_feature_importance.loc[random_forest_feature_importance.importance<0.01].variable.values[-i]}")
bottom_variable = random_forest_feature_importance.loc[random_forest_feature_importance.importance<0.01].variable.values[-i]
bottom_variable = bottom_variable.split('_')[0]
bottom_variable = [col for col in list(x) if bottom_variable in col]
compare_counter = 0
for var in bottom_variable:
if var in random_forest_feature_importance.loc[random_forest_feature_importance.importance<0.01].variable.values:
compare_counter += 1
if len(bottom_variable) == compare_counter:
print(f"Following variable(s) will be dropped from x {bottom_variable}")
x = x.drop(columns=bottom_variable)
break
else:
print("Next 'worst' variable will be examined for dropping.")
continue
else:
break
# Create DataFrame of random forest classifier results
model_search_rfc = pd.DataFrame(model_search_rfc, columns=['best_model_params_grid_search', 'best_score_grid_search',
'true_negatives', 'false_positives',
'false_negatives', 'true_positives', 'variables_not_used'])
# Create recall and precision columns
model_search_rfc['recall'] = model_search_rfc.true_positives/(model_search_rfc.true_positives + model_search_rfc.false_negatives)
model_search_rfc['precision'] = model_search_rfc.true_positives/(model_search_rfc.true_positives + model_search_rfc.false_positives)
model_search_rfc['f1_score'] = 2 * (model_search_rfc.precision * model_search_rfc.recall) / (model_search_rfc.precision + model_search_rfc.recall)
# Sort DataFrame
model_search_rfc = model_search_rfc.sort_values(by=['f1_score'], ascending=False)
print(model_search_rfc)
if len(model_search_rfc.loc[model_search_rfc.f1_score==model_search_rfc.f1_score.max()]) > 1:
top_model_result_rfc = model_search_rfc.loc[(model_search_rfc.f1_score==model_search_rfc.f1_score.max()) &
(model_search_rfc['variables_not_used'].apply(len) == max(map(lambda x: len(x[list(model_search_rfc).index('variables_not_used')]),
model_search_rfc.loc[model_search_rfc.f1_score==model_search_rfc.f1_score.max()].values)))]
if len(top_model_result_rfc) > 1:
print("Fix multiple best model problem for rfc")
break
else:
top_model_result_rfc = model_search_rfc.loc[model_search_rfc.f1_score==model_search_rfc.f1_score.max()]
top_model_results = top_model_results.append(other=top_model_result_rfc, sort=False)
print(f"Top rfc model: \n {top_model_result_rfc}")
# Append top_model_result_rfc results to all_model_results DataFrame
# Re-create feature variables
x = model.drop(columns='num')
rfc_predict_proba = cross_val_predict(RandomForestClassifier(criterion=top_model_result_rfc["best_model_params_grid_search"].values[0]['criterion'],
max_features=top_model_result_rfc["best_model_params_grid_search"].values[0]['max_features'],
n_estimators=top_model_result_rfc["best_model_params_grid_search"].values[0]['n_estimators']),
x[[x for x in list(x) if x not in list(top_model_result_rfc['variables_not_used'].values[0])]], y,
cv=5, method='predict_proba')
all_model_results['rfc_'+inflect.engine().number_to_words(index)+'_pred_zero'] = rfc_predict_proba[:,0]
all_model_results['rfc_'+inflect.engine().number_to_words(index)+'_pred_one'] = rfc_predict_proba[:,1]
# Fill in model_type columns
top_model_results['model_type'] = top_model_results['model_type'].fillna(value='rfc')
Support Vector Machine
# Unique variable combination runs
for index, (vars_to_drop, cat_vars_to_model) in enumerate(zip(variables_to_drop_list,
categorical_variables_for_modeling_list), start=1):
print(f"Model run: {index}")
# Create copy of hungarian for non-regression modeling
model = hungarian.copy()
# Drop columns
model = model.drop(columns=vars_to_drop)
# Dummy variable categorical variables
model = pd.get_dummies(data=model, columns=cat_vars_to_model, drop_first=False)
# Create target variable
y = model['num']
# Create feature variables
x = model.drop(columns='num')
# Create copy of x for standard scaling
x_std = x.copy()
# print(list(x_std)[list(x_std).index('sex_0')-1])
x_std.loc[:, :list(x_std)[list(x_std).index('sex_0')-1]] = scaler.fit_transform(x_std.loc[:, :list(x_std)[list(x_std).index('sex_0')-1]])
# Define parameters of SVC
svc_model = SVC(kernel='linear')
# Recursive feature elimination
rfe_svc = pd.DataFrame(data=[list(x_std), RFE(svc_model, n_features_to_select=1).fit(x_std, y).ranking_.tolist()]).T.\
rename(columns={0: 'variable', 1: 'rfe_ranking'}).sort_values(by='rfe_ranking').reset_index(drop=True)
svc_model = SVC(random_state=1)
param_grid = {'kernel': ['rbf', 'sigmoid', 'linear'], 'C': np.arange(0.10, 2.41, step=0.05), 'gamma': ['scale', 'auto']}
cv = ShuffleSplit(n_splits=5, test_size=0.3)
# Define grid search CV parameters
grid_search = GridSearchCV(svc_model, param_grid, cv=cv)
# Loop through features based on recursive feature elimination evaluation - top to bottom
model_search_svc = []
svc_variable_list = []
for i in range(len(rfe_svc)):
if rfe_svc['variable'][i] not in svc_variable_list:
svc_variable_list.extend([rfe_svc['variable'][i]])
# Add related one-hot encoded variables if variable is categorical
if svc_variable_list[-1].split('_')[-1] in sorted([x for x in list(set([x.split('_')[-1] for x in list(x_std)])) if len(x) == 1]):
svc_variable_list.extend([var for var in list(x_std) if svc_variable_list[-1].split('_')[0] in var and var != svc_variable_list[-1]])
print(svc_variable_list)
grid_search.fit(x_std[svc_variable_list], y)
print(f'Best parameters for current grid seach: {grid_search.best_params_}')
print(f'Best score for current grid seach: {grid_search.best_score_}')
# Define parameters of Support-vector machine classifier from grid search
svc_model = SVC(kernel=grid_search.best_params_['kernel'], C=grid_search.best_params_['C'],
gamma=grid_search.best_params_['gamma'], random_state=1)
# Cross-validate and predict using Support-vector machine classifier
svc_predict = cross_val_predict(svc_model, x_std[svc_variable_list], y, cv=5)
print(confusion_matrix(y_true=y, y_pred=svc_predict))
conf_matr = confusion_matrix(y_true=y, y_pred=svc_predict)
model_search_svc.append([grid_search.best_params_, grid_search.best_score_, conf_matr[0][0],
conf_matr[0][1], conf_matr[1][0], conf_matr[1][1], list(x_std[svc_variable_list])])
# Create DataFrame of svc results
model_search_svc = pd.DataFrame(model_search_svc, columns=['best_model_params_grid_search', 'best_score_grid_search',
'true_negatives', 'false_positives',
'false_negatives', 'true_positives', 'variables_used'])
# Create recall, precision, f1-score columns
model_search_svc['recall'] = model_search_svc.true_positives/(model_search_svc.true_positives + model_search_svc.false_negatives)
model_search_svc['precision'] = model_search_svc.true_positives/(model_search_svc.true_positives + model_search_svc.false_positives)
model_search_svc['f1_score'] = 2 * (model_search_svc.precision * model_search_svc.recall) / (model_search_svc.precision + model_search_svc.recall)
# Sort DataFrame
model_search_svc = model_search_svc.sort_values(by=['f1_score'], ascending=False)
print(model_search_svc)
# Choose top model from svc model search
if len(model_search_svc.loc[model_search_svc.f1_score==model_search_svc.f1_score.max()]) > 1:
top_model_result_svc = model_search_svc.loc[(model_search_svc.f1_score == model_search_svc.f1_score.max()) &
(model_search_svc['variables_used'].apply(len) == min(map(lambda x: len(x[list(model_search_svc).index('variables_used')]),
model_search_svc.loc[model_search_svc.f1_score==model_search_svc.f1_score.max()].values)))]
if len(top_model_result_svc) > 1:
print('break here')
break
# top_model_result_svc = top_model_result_svc.loc[top_model_result_svc.best_score_grid_search == top_model_result_svc.best_score_grid_search.max()]
else:
top_model_result_svc = model_search_svc.loc[model_search_svc.f1_score==model_search_svc.f1_score.max()]
top_model_results = top_model_results.append(other=top_model_result_svc, sort=False)
print(f"Top svc model: \n {top_model_result_svc}")
# Append top_model_result_svc results to all_model_results DataFrame
all_model_results['svc_'+inflect.engine().number_to_words(index)] = cross_val_predict(SVC(
kernel=top_model_result_svc['best_model_params_grid_search'].values[0]['kernel'],
C=top_model_result_svc['best_model_params_grid_search'].values[0]['C'],
gamma=top_model_result_svc['best_model_params_grid_search'].values[0]['gamma']),
x_std[top_model_result_svc['variables_used'].values[0]], y, cv=5)
# Fill in model_type columns
top_model_results['model_type'] = top_model_results['model_type'].fillna(value='svc')
K-Nearest Neighbors
# Unique variable combination runs
for index, (vars_to_drop, cat_vars_to_model) in enumerate(zip(variables_to_drop_list,
categorical_variables_for_modeling_list), start=1):
print(f"Model run: {index}")
# Create copy of hungarian for non-regression modeling
model = hungarian.copy()
# Drop columns
model = model.drop(columns=vars_to_drop)
# Dummy variable categorical variables
model = pd.get_dummies(data=model, columns=cat_vars_to_model, drop_first=False)
# Create target variable
y = model['num']
# Create feature variables
x = model.drop(columns='num')
# Create copy of x for standard scaling
x_std = x.copy()
x_std.loc[:, :list(x_std)[list(x_std).index('sex_0')-1]] = scaler.fit_transform(x_std.loc[:, :list(x_std)[list(x_std).index('sex_0')-1]])
# Use Recursive Feature Elimination from SVC
# Define parameters of SVC
svc_model = SVC(kernel='linear', random_state=1)
# Feature importance DataFrame
feature_info = pd.DataFrame(data=[list(x_std), RFE(svc_model, n_features_to_select=1).fit(x_std, y).ranking_.tolist()]).T.\
rename(columns={0: 'variable', 1: 'rfe_svc'}).reset_index(drop=True)
# Define parameters of Random Forest Classifier
random_forest_model = RandomForestClassifier(random_state=1)
# Merge feature importances from random forest classifier on feature_info
feature_info = feature_info.merge(pd.DataFrame(data=[list(x), random_forest_model.fit(x,y).feature_importances_.tolist()]).T.\
rename(columns={0: 'variable', 1: 'feature_importance_rfc'}), on='variable')
# Sort values by descending random forest classifier feature importance to create ranking column
feature_info = feature_info.sort_values(by='feature_importance_rfc', ascending=False)
feature_info['feature_importance_rfc_ranking'] = np.arange(1,len(feature_info)+1)
# Define parameters of Gradient Boosting Classifier
gbm_model = GradientBoostingClassifier(random_state=1)
# Merge feature importances from gradient boosting classifier on feature_info
feature_info = feature_info.merge(pd.DataFrame(data=[list(x), gbm_model.fit(x,y).feature_importances_.tolist()]).T.\
rename(columns={0: 'variable', 1: 'feature_importance_gbm'}), on='variable')
# Sort values by descending gradient boosting classifier feature importance to create ranking column
feature_info = feature_info.sort_values(by='feature_importance_gbm', ascending=False)
feature_info['feature_importance_gbm_ranking'] = np.arange(1,len(feature_info)+1)
# Get average of three RFE/feature importance columns
feature_info['feature_importance_avg'] = feature_info[['rfe_svc', 'feature_importance_rfc_ranking', 'feature_importance_gbm_ranking']].mean(axis=1)
# Sort values by average column
feature_info = feature_info.sort_values(by='feature_importance_avg', ascending=True).reset_index(drop=True)
# Define parameters of kNN model
knn_model = KNeighborsClassifier(metric='minkowski')
# Define parameters of grid search
param_grid = {'n_neighbors': np.arange(9, 47, step=2), 'weights': ['uniform', 'distance']}
# Define parameters of shuffle split
cv = ShuffleSplit(n_splits=5, test_size=0.3)
# Define grid search CV parameters
grid_search = GridSearchCV(knn_model, param_grid, cv=cv) # , scoring='recall'
# Append model results to this list
model_search_knn = []
# Begin top to bottom process - looking at most important variables (by RFE ranking first and adding on)
knn_variable_list = []
for i in range(len(feature_info)):
if feature_info['variable'][i] not in knn_variable_list:
knn_variable_list.extend([feature_info['variable'][i]])
# Add related one-hot encoded variables if variable is categorical
if knn_variable_list[-1].split('_')[-1] in sorted([x for x in list(set([x.split('_')[-1] for x in list(x_std)])) if len(x) == 1]):
knn_variable_list.extend([var for var in list(x_std) if knn_variable_list[-1].split('_')[0] in var and var != knn_variable_list[-1]])
print(knn_variable_list)
grid_search.fit(x_std[knn_variable_list], y)
print(f'Best parameters for current grid seach: {grid_search.best_params_}')
print(f'Best score for current grid seach: {grid_search.best_score_}')
# Define parameters of k-nearest neighbors from grid search
knn_model = KNeighborsClassifier(metric='minkowski', n_neighbors=grid_search.best_params_['n_neighbors'],
weights=grid_search.best_params_['weights'])
# Cross-validate and predict using Support-vector machine classifier
knn_predict = cross_val_predict(knn_model, x_std[knn_variable_list], y, cv=5)
print(confusion_matrix(y_true=y, y_pred=knn_predict))
conf_matr = confusion_matrix(y_true=y, y_pred=knn_predict)
model_search_knn.append([grid_search.best_params_, grid_search.best_score_, conf_matr[0][0],
conf_matr[0][1], conf_matr[1][0], conf_matr[1][1], list(x_std[knn_variable_list])])
# Create DataFrame of k-nearest neighbors results
model_search_knn = pd.DataFrame(model_search_knn, columns=['best_model_params_grid_search', 'best_score_grid_search',
'true_negatives', 'false_positives',
'false_negatives', 'true_positives', 'variables_used'])
# Create recall, precision, f1-score columns
model_search_knn['recall'] = model_search_knn.true_positives/(model_search_knn.true_positives + model_search_knn.false_negatives)
model_search_knn['precision'] = model_search_knn.true_positives/(model_search_knn.true_positives + model_search_knn.false_positives)
model_search_knn['f1_score'] = 2 * (model_search_knn.precision * model_search_knn.recall) / (model_search_knn.precision + model_search_knn.recall)
# Sort DataFrame
model_search_knn = model_search_knn.sort_values(by=['f1_score'], ascending=False)
print(model_search_knn)
if len(model_search_knn.loc[model_search_knn.f1_score==model_search_knn.f1_score.max()]) > 1:
print("Fix multiple best model problem for rfc")
break
else:
top_model_result_knn = model_search_knn.loc[model_search_knn.f1_score==model_search_knn.f1_score.max()]
top_model_results = top_model_results.append(other=top_model_result_knn, sort=False)
print(f"Top knn model: \n {top_model_result_knn}")
# Append top_model_result_knn results to all_model_results DataFrame
knn_predict_proba = cross_val_predict(KNeighborsClassifier(metric='minkowski',
n_neighbors=top_model_result_knn['best_model_params_grid_search'].values[0]['n_neighbors'],
weights=top_model_result_knn['best_model_params_grid_search'].values[0]['weights']),
x_std[top_model_result_knn['variables_used'].values[0]], y, cv=5, method='predict_proba')
all_model_results['knn_'+inflect.engine().number_to_words(index)+'_pred_zero'] = knn_predict_proba[:,0]
all_model_results['knn_'+inflect.engine().number_to_words(index)+'_pred_one'] = knn_predict_proba[:,1]
# Fill in model_type columns
top_model_results['model_type'] = top_model_results['model_type'].fillna(value='knn')
Gradient Boosting
# Unique variable combination runs
for index, (vars_to_drop, cat_vars_to_model) in enumerate(zip(variables_to_drop_list,
categorical_variables_for_modeling_list), start=1):
print(f"Model run: {index}")
# Create copy of hungarian for non-regression modeling
model = hungarian.copy()
# Drop columns
model = model.drop(columns=vars_to_drop)
# Dummy variable categorical variables
model = pd.get_dummies(data=model, columns=cat_vars_to_model, drop_first=False)
# Create target variable
y = model['num']
# Create feature variables
x = model.drop(columns='num')
# Obtain list of all feature variables
x_all = list(x)
model_search_gbm = []
while True:
print(x.shape)
print('------')
print("\n")
# Baseline model
gbm_baseline = GradientBoostingClassifier()
# Cross-validate
cross_val_score_gbm = cross_val_score(gbm_baseline, x, y, cv=cv)
# Begin parameter tuning for GBM
# Set initial values (will be tuned later)
min_samples_split = 3
min_samples_leaf = 20
max_depth = 5
max_features = 'sqrt'
subsample = 0.8
learning_rate = 0.1
# Set param_grid to tune n_estimators
param_grid = {'n_estimators': np.arange(20,81,10)}
# Tune n_estimators
gbm_one = GradientBoostingClassifier(learning_rate= learning_rate, min_samples_split= min_samples_split,
min_samples_leaf= min_samples_leaf, max_depth= max_depth,
max_features= max_features, subsample= subsample)
grid_search = GridSearchCV(gbm_one, param_grid, cv=cv) # , scoring='recall'
grid_search.fit(x, y)
# Obain n_estimators from grid search
n_estimators_best_param_grid_search_one = grid_search.best_params_['n_estimators']
# Tune tree-specific parameters
param_grid2 = {'max_depth': np.arange(3,20,2), 'min_samples_split': np.arange(10,200,10)}
# Tune max_depth and min_samples_split
gbm_two = GradientBoostingClassifier(learning_rate= learning_rate, max_features= max_features,
subsample= subsample, n_estimators=n_estimators_best_param_grid_search_one)
grid_search = GridSearchCV(gbm_two, param_grid2, cv=cv) # , scoring='recall'
grid_search.fit(x, y)
# Obain max_depth and min_samples_split from grid search
max_depth_best_param_grid_search_two = grid_search.best_params_['max_depth']
min_samples_split_best_param_grid_search_two = grid_search.best_params_['min_samples_split']
# Tune min_samples_leaf
param_grid3 = {'min_samples_leaf': np.arange(1,15,1)}
gbm_three = GradientBoostingClassifier(learning_rate= learning_rate, max_features= max_features,
subsample= subsample, n_estimators=n_estimators_best_param_grid_search_one,
max_depth= max_depth_best_param_grid_search_two,
min_samples_split= min_samples_split_best_param_grid_search_two)
grid_search = GridSearchCV(gbm_three, param_grid3, cv=cv) # , scoring='recall'
grid_search.fit(x, y)
# Obain min_samples_leaf from grid search
min_samples_leaf_best_param_grid_search_three = grid_search.best_params_['min_samples_leaf']
# Tune max_features
param_grid4 = {'max_features': np.arange(2,20,1)}
gbm_four = GradientBoostingClassifier(learning_rate= learning_rate, subsample= subsample,
n_estimators=n_estimators_best_param_grid_search_one,
max_depth= max_depth_best_param_grid_search_two,
min_samples_split= min_samples_split_best_param_grid_search_two,
min_samples_leaf= min_samples_leaf_best_param_grid_search_three)
grid_search = GridSearchCV(gbm_four, param_grid4, cv=cv) # , scoring='recall'
grid_search.fit(x, y)
# Obain max_features from grid search
max_features_best_param_grid_search_four = grid_search.best_params_['max_features']
# Tune subsample
param_grid5 = {'subsample': np.arange(0.6,1,0.05)}
gbm_five = GradientBoostingClassifier(learning_rate= learning_rate,
n_estimators=n_estimators_best_param_grid_search_one,
max_depth=max_depth_best_param_grid_search_two,
min_samples_split=min_samples_split_best_param_grid_search_two,
min_samples_leaf=min_samples_leaf_best_param_grid_search_three,
max_features=max_features_best_param_grid_search_four)
grid_search = GridSearchCV(gbm_five, param_grid5, cv=cv) # , scoring='recall'
grid_search.fit(x, y)
# Obtain subsample from grid search
subsample_best_param_grid_search_five = grid_search.best_params_['subsample']
# Tune learning rate and increase n_estimators proportionally
param_grid_list = [[learning_rate, n_estimators_best_param_grid_search_one],
[learning_rate/2, n_estimators_best_param_grid_search_one*2],
[learning_rate/5, n_estimators_best_param_grid_search_one*5],
[learning_rate/10, n_estimators_best_param_grid_search_one*10],
[learning_rate/20, n_estimators_best_param_grid_search_one*20],
[learning_rate/30, n_estimators_best_param_grid_search_one*30],
[learning_rate/40, n_estimators_best_param_grid_search_one*40],
[learning_rate/50, n_estimators_best_param_grid_search_one*50]]
# Append l_rate, n_ests, and cross_val_score mean
cross_val_score_gbm_six_means = []
for l_rate, n_ests in param_grid_list:
gbm_six = GradientBoostingClassifier(learning_rate=l_rate, n_estimators=n_ests,
max_depth=max_depth_best_param_grid_search_two,
min_samples_split=min_samples_split_best_param_grid_search_two,
min_samples_leaf=min_samples_leaf_best_param_grid_search_three,
max_features=max_features_best_param_grid_search_four,
subsample=subsample_best_param_grid_search_five)
cross_val_score_gbm_six = cross_val_score(gbm_six, x, y, cv=cv)
cross_val_score_gbm_six_means.append([l_rate, n_ests, cross_val_score_gbm_six.mean()])
# Retrieve best values for learning_rate and n_estimators based on max value of cross_val_score_gbm_six.mean()
learning_rate_n_estimators_best_param_grid_list = list(filter(lambda x: x[2] == max(map(lambda x: x[2],
cross_val_score_gbm_six_means)), cross_val_score_gbm_six_means))[0]
gbm_final = GradientBoostingClassifier(learning_rate=learning_rate_n_estimators_best_param_grid_list[0],
n_estimators=learning_rate_n_estimators_best_param_grid_list[1],
max_depth=max_depth_best_param_grid_search_two,
min_samples_split=min_samples_split_best_param_grid_search_two,
min_samples_leaf=min_samples_leaf_best_param_grid_search_three,
max_features=max_features_best_param_grid_search_four,
subsample=subsample_best_param_grid_search_five)
gbm_predict = cross_val_predict(gbm_final, x, y, cv=5)
conf_matr = confusion_matrix(y_true=y, y_pred=gbm_predict)
model_search_gbm.append([gbm_final.get_params(), conf_matr[0][0], conf_matr[0][1],
conf_matr[1][0], conf_matr[1][1], set(x_all).difference(x)])
# Obtain feature importances
gradient_boosting_feature_importance = pd.DataFrame(data=[list(x),
gbm_final.fit(x, y).feature_importances_.tolist()]).T.rename(columns={0:'variable',
1:'importance'}).sort_values(by='importance', ascending=False)
# Remove 'worst' variables one-by-one
print(gradient_boosting_feature_importance.loc[gradient_boosting_feature_importance.importance < 0.10])
if len(gradient_boosting_feature_importance.loc[gradient_boosting_feature_importance.importance < 0.01]) > 0:
for i in range(1, len(gradient_boosting_feature_importance.loc[gradient_boosting_feature_importance.importance < 0.01]) + 1):
print(f"'Worst' variable being examined: "
f"{gradient_boosting_feature_importance.loc[gradient_boosting_feature_importance.importance < 0.01].variable.values[-i]}")
bottom_variable = gradient_boosting_feature_importance.loc[gradient_boosting_feature_importance.importance < 0.01].variable.values[-i]
# Add related one-hot encoded variables if variable is categorical
if bottom_variable.split('_')[-1] in sorted([x for x in list(set([x.split('_')[-1] for x in list(x)])) if len(x) == 1]):
bottom_variable = bottom_variable.split('_')[0]
bottom_variable = [col for col in list(x) if bottom_variable in col]
compare_counter = 0
for var in bottom_variable:
if var in gradient_boosting_feature_importance.loc[gradient_boosting_feature_importance.importance<0.01].variable.values:
compare_counter += 1
if len(bottom_variable) == compare_counter:
print(f"Following variable(s) will be dropped from x {bottom_variable}")
x = x.drop(columns=bottom_variable)
break
else:
print("Next 'worst' variable will be examined for dropping.")
continue
# else:
# x = x.drop(columns=bottom_variable)
else:
break
# Create DataFrame of random forest classifier results
model_search_gbm = pd.DataFrame(model_search_gbm, columns=['model_params_grid_search',
'true_negatives', 'false_positives',
'false_negatives', 'true_positives', 'variables_not_used'])
# Create recall and precision columns
model_search_gbm['recall'] = model_search_gbm.true_positives/(model_search_gbm.true_positives + model_search_gbm.false_negatives)
model_search_gbm['precision'] = model_search_gbm.true_positives/(model_search_gbm.true_positives + model_search_gbm.false_positives)
model_search_gbm['f1_score'] = 2 * (model_search_gbm.precision * model_search_gbm.recall) / (model_search_gbm.precision + model_search_gbm.recall)
# Sort DataFrame
model_search_gbm = model_search_gbm.sort_values(by=['f1_score'], ascending=False)
print(model_search_gbm)
# Choose top model from gbm model search
if len(model_search_gbm.loc[model_search_gbm.f1_score==model_search_gbm.f1_score.max()]) > 1:
top_model_result_gbm = model_search_gbm.loc[(model_search_gbm.f1_score == model_search_gbm.f1_score.max()) &
(model_search_gbm['variables_not_used'].apply(len) == max(map(lambda x: len(x[list(model_search_gbm).index('variables_not_used')]),
model_search_gbm.loc[model_search_gbm.f1_score==model_search_gbm.f1_score.max()].values)))]
else:
top_model_result_gbm = model_search_gbm.loc[model_search_gbm.f1_score == model_search_gbm.f1_score.max()]
top_model_results = top_model_results.append(other=top_model_result_gbm, sort=False)
print(f"Top gbm model: \n {top_model_result_gbm}")
# Append top_model_result_rfc results to all_model_results DataFrame
# Re-create feature variables
x = model.drop(columns='num')
gbm_predict_proba = cross_val_predict(GradientBoostingClassifier(learning_rate=top_model_result_gbm["model_params_grid_search"].values[0]["learning_rate"],
n_estimators=top_model_result_gbm["model_params_grid_search"].values[0]["n_estimators"],
max_depth=top_model_result_gbm["model_params_grid_search"].values[0]["max_depth"],
min_samples_split=top_model_result_gbm["model_params_grid_search"].values[0]["min_samples_split"],
min_samples_leaf=top_model_result_gbm["model_params_grid_search"].values[0]["min_samples_leaf"],
max_features=top_model_result_gbm["model_params_grid_search"].values[0]["max_features"],
subsample=top_model_result_gbm["model_params_grid_search"].values[0]["subsample"]),
x[[x for x in list(x) if x not in list(top_model_result_gbm['variables_not_used'].values[0])]], y, cv=5, method='predict_proba')
all_model_results['gbm_'+inflect.engine().number_to_words(index)+'_pred_zero'] = gbm_predict_proba[:,0]
all_model_results['gbm_'+inflect.engine().number_to_words(index)+'_pred_one'] = gbm_predict_proba[:,1]
# Fill in model_type columns
top_model_results['model_type'] = top_model_results['model_type'].fillna(value='gbm')
Save Created Model Results DataFrames
# Save top_model_results to csv
top_model_results.to_pickle('top_model_results.pkl')
# Save all_model_results to csv
all_model_results.to_pickle('all_model_results.pkl')
Model Visualization Comparison and Selection
Load in Model Results DataFrames if Needed
# Check if DataFrame is already loaded in - if not, load from pickle file
try:
top_model_results
except NameError:
with open('top_model_results.pkl', 'rb') as top_model_results_pkl:
top_model_results = pickle.load(top_model_results_pkl)
# Check if DataFrame is already loaded in - if not, load from pickle file
try:
all_model_results
except NameError:
with open('all_model_results.pkl', 'rb') as all_model_results_pkl:
all_model_results = pickle.load(all_model_results_pkl)
# Check if target variable is defined as y - if not, load from pickle file
try:
y
except NameError:
with open('hungarian_target_variable.pkl', 'rb') as hungarian_target_variable_pkl:
y = pickle.load(hungarian_target_variable_pkl)
ROC Curves
# Dict of model names and their spelled out verions
model_names_spelled_out = {'logit': 'Logistic Regression', 'rfc': 'Random Forest Classifier', 'knn': 'K-Nearest Neighbors',
'svc': 'Support Vector Machine Classifier', 'gbm': 'Gradient Boosting Classifier'}
# Build ROC Curves for all models which give prediction probabilities (i.e. all but SVC)
fig, axes = plt.subplots(nrows=2, ncols=2)
fig.subplots_adjust(left=0.05, right=0.87, top=0.90, bottom=0.07, hspace=0.35, wspace=0.20)
fig.suptitle('ROC (Receiver Operating Characteristic) Curves', fontweight= 'bold', fontsize= 22)
for ax, value in zip(axes.flatten(), list(unique_everseen([x.split("_")[0] for x in all_model_results.columns if 'pred' in x]))):
# ROC Curve plot
for pred_one_col in [x for x in all_model_results.columns if (x[0:len(value)] == value) & (x[-len('pred_one'):] == 'pred_one')]:
fpr, tpr, thresholds = roc_curve(y, all_model_results[pred_one_col])
ax.plot(fpr, tpr, label=pred_one_col.split("_")[1])
ax.plot([0, 1], [0, 1],'r--')
ax.set_xlim([0.0, 1.0])
ax.set_ylim([0.0, 1.05])
ax.set_xlabel('False Positive Rate', fontdict={'fontweight': 'bold', 'fontsize': 18}, labelpad=15)
ax.set_ylabel('True Positive Rate', fontdict={'fontweight': 'bold', 'fontsize': 18}, labelpad=15)
ax.set_title(f'{model_names_spelled_out[value]} Models', fontdict={'fontweight': 'bold', 'fontsize': 24})
handles, legends = ax.get_legend_handles_labels()
legends = ['Model ' + legend.title() for legend in legends]
fig.legend(handles, legends, loc='upper left', bbox_to_anchor=(0.87, 0.85), prop={'weight':'bold', 'size':16})
# Expand figure to desired size first before running below code (this makes xtick and ytick labels appear and then can therefore be bolded)
for ax in axes.flatten():
ax.set_yticklabels(labels=ax.get_yticklabels(), fontweight="bold")
ax.set_xticklabels(labels=ax.get_xticklabels(), fontweight="bold")
Select Best Model for Each Algorithm
# Re-assign index of top_model_results
top_model_results.index = list(itertools.chain.from_iterable(itertools.repeat(range(1,8), 5)))
# Retreive best model run from each algorithm (base off f-1 score)
top_model_from_each_algorithm = pd.DataFrame(columns=top_model_results.columns)
for value in list(unique_everseen(top_model_results.model_type)):
if len(top_model_results.loc[(top_model_results.model_type == value) &
(top_model_results.f1_score == top_model_results.loc[(top_model_results.model_type == value), 'f1_score'].max())]) > 1:
# Create DataFrame to get best model from remaining
gt_one_top_model = top_model_results.loc[(top_model_results.model_type == value) &
(top_model_results.f1_score == top_model_results.loc[(top_model_results.model_type == value), 'f1_score'].max())]
if value == 'logit':
# Get DataFrame with least amount of variables used (parsimonious model)
if len(gt_one_top_model.loc[gt_one_top_model.variables_used.apply(len) == gt_one_top_model.variables_used.apply(len).min()])>1:
# Sample random one from remaining
top_model_from_each_algorithm = top_model_from_each_algorithm.append(other=gt_one_top_model.sample(n=1))
else:
print(value)
break
else:
top_model_from_each_algorithm = top_model_from_each_algorithm.append(other=top_model_results.loc[(top_model_results.model_type == value) &
(top_model_results.f1_score == top_model_results.loc[(top_model_results.model_type == value), 'f1_score'].max())])
all_cut_offs = []
# Go through various cut-offs for each model run that returns predicted probabilities
for value in list(unique_everseen([x.split("_")[0] for x in all_model_results.columns if 'pred' in x])):
print("-" * 80)
print(value)
for pred_one_col in [x for x in all_model_results.columns if (x[0:len(value)] == value) & (x[-len('pred_one'):] == 'pred_one')]:
print(pred_one_col)
for cut_off in np.arange(0,1.01,step=0.001):
print(cut_off)
print("-" * 80)
cf = confusion_matrix(y_true=y, y_pred=np.where(all_model_results[pred_one_col] > cut_off, 1, 0))
all_cut_offs.append([pred_one_col, cut_off, cf[0][0], cf[0][1], cf[1][0], cf[1][1]])
# Create DataFrame of results
all_cut_offs = pd.DataFrame(all_cut_offs, columns=["model", "cut_off", 'true_negatives', 'false_positives',
'false_negatives', 'true_positives'])
# Create recall, precision, and f1_score columns
all_cut_offs['recall'] = all_cut_offs.true_positives/(all_cut_offs.true_positives + all_cut_offs.false_negatives)
all_cut_offs['precision'] = all_cut_offs.true_positives/(all_cut_offs.true_positives + all_cut_offs.false_positives)
all_cut_offs['f1_score'] = 2 * (all_cut_offs.precision * all_cut_offs.recall) / (all_cut_offs.precision + all_cut_offs.recall)
# Create total correct and total wrong columns
all_cut_offs["total_correct"] = all_cut_offs.true_negatives + all_cut_offs.true_positives
all_cut_offs["total_wrong"] = all_cut_offs.false_positives + all_cut_offs.false_negatives
# Groupby and take top row from each groupby
all_cut_offs = all_cut_offs.groupby(["model"]).apply(lambda x: (x.sort_values(["total_correct",
"f1_score"], ascending = [False, False])).head(1)).reset_index(drop=True)
# Get the model type (first part of model before "_") to groupby on it
all_cut_offs["model_type"] = all_cut_offs["model"].apply(lambda x: x.split("_")[0])
# Get top model from each algorithm
all_cut_offs = all_cut_offs.groupby(["model_type"]).apply(lambda x: (x.sort_values(["total_correct",
"f1_score"], ascending = [False, False])).head(1)).reset_index(drop=True)
all_cut_offs_results = pd.DataFrame()
# Get predictions with each of the models
for model, cut_off in zip(all_cut_offs.model, all_cut_offs.cut_off):
print(model, cut_off)
all_cut_offs_results[model] = np.where(all_model_results[model] > cut_off, 1, 0)
# Append best svc model to all_cut_offs_results
all_cut_offs_results["svc_" + inflect.engine().number_to_words(
top_model_from_each_algorithm.loc[top_model_from_each_algorithm.model_type=="svc"].index[0])] = \
all_model_results["svc_" + inflect.engine().number_to_words(
top_model_from_each_algorithm.loc[top_model_from_each_algorithm.model_type=="svc"].index[0])]
Determine Combination of Models or Stand Alone Model Which Provides Best Results
# Get combinations of length 1,3, and 5 (take mode of 3 and 5)
model_search_all = []
for length in [1, 3, 5]:
for comb in itertools.combinations(all_cut_offs_results.columns, length):
print(list(comb))
print(confusion_matrix(y_true=y, y_pred=all_cut_offs_results[list(comb)].mode(axis=1)))
conf_matr = confusion_matrix(y_true=y, y_pred=all_cut_offs_results[list(comb)].mode(axis=1))
model_search_all.append([comb, conf_matr[0][0], conf_matr[0][1], conf_matr[1][0], conf_matr[1][1]])
print('\n')
# Create DataFrame of results
model_search_all = pd.DataFrame(model_search_all, columns=['cols', 'true_negatives', 'false_positives',
'false_negatives', 'true_positives'])
# Create recall, precision, and f1-score columns
model_search_all['recall'] = model_search_all.true_positives/(model_search_all.true_positives + model_search_all.false_negatives)
model_search_all['precision'] = model_search_all.true_positives/(model_search_all.true_positives + model_search_all.false_positives)
model_search_all['f1_score'] = 2 * (model_search_all.precision * model_search_all.recall) / (model_search_all.precision + model_search_all.recall)
# Create total correct and total wrong columns
model_search_all["total_correct"] = model_search_all.true_negatives + model_search_all.true_positives
model_search_all["total_wrong"] = model_search_all.false_positives + model_search_all.false_negatives
# Sort DataFrame
model_search_all = model_search_all.sort_values(by=['total_correct','f1_score'], ascending=[False, False])
print(model_search_all)
Create Model Results Summary Table for Display
# Spell out all model names - return list of lists
column_all = []
for index, value in enumerate(model_search_all['cols']):
one_column = []
for cols in model_search_all['cols'].iloc[index]:
if cols.split("_")[0] in model_names_spelled_out.keys():
cols_split = cols.split("_")
cols_split[0] = model_names_spelled_out[cols_split[0]]
cols_split = "_".join(cols_split)
cols_split = cols_split.replace("pred_one", "").replace("_", " ").title().strip()
one_column.extend([cols_split])
column_all.append(one_column)
# Convert each list to a string and add as columns
model_search_all['columns'] = [', '.join(map(str, l)) for l in column_all]
# Create table for display
model_search_all[['columns', 'f1_score', 'recall', 'precision', 'total_correct', 'total_wrong']].rename(columns={'columns': 'Model(s)',
'f1_score': 'F1 Score',
'recall': 'Recall', 'precision': 'Precision',
'total_correct': 'Total Correct',
'total_wrong': 'Total Incorrect'}).to_csv("final_models_table.csv", index=False)
Go Back to Model Visualization, Comparison, and Selection
Visualize Best Model
Confusion Matrix of Best Model
conf_matrix = [model_search_all.loc[model_search_all.cols==('svc_four',)][["true_negatives",
"false_positives"]].values[0].tolist(),
model_search_all.loc[model_search_all.cols==('svc_four',)][["false_negatives",
"true_positives"]].values[0].tolist()]
group_names = ['Correctly Predicted To\nNot Have Heart Disease\n', 'Incorrectly Predicted To\nHave Heart Disease\n',
'Incorrectly Predicted To\nNot Have Heart Disease\n', 'Correctly Predicted To\nHave Heart Disease\n']
group_counts = conf_matrix[0] + conf_matrix[1]
labels = [f"{v1}\n{v2}" for v1, v2 in zip(group_names, group_counts)]
labels = np.asarray(labels).reshape(2, 2)
tick_labels = ['No Presence of Heart Disease', 'Presence of Heart Disease']
# Set figsize to size of second monitor
plt.rcParams['figure.figsize'] = [19.2, 9.99]
fig, axes = plt.subplots(nrows=1, ncols=1)
fig.subplots_adjust(left=0.07, right=0.99, top=0.94, bottom=0.11)
sns.heatmap(conf_matrix, annot=labels, annot_kws={"size": 26, "weight": "bold"}, fmt='', xticklabels=tick_labels,
yticklabels=tick_labels, cbar=False, cmap=['#b2abd2', '#e66101'],
center=model_search_all.loc[model_search_all.cols==('svc_four',)][["false_positives", "false_negatives"]].values.max())
plt.xticks(weight="bold", size=20)
plt.yticks(rotation=90, va='center', weight="bold", size=20)
plt.ylabel('True Value of Patient', weight="bold", size=24, labelpad=30)
plt.xlabel('Predicted Value of Patient', weight="bold", size=24, labelpad=30)
plt.title('Prediction Results for Support Vector Machine Classifier Model #4', pad=15, fontdict={"weight": "bold", "size": 28})
plt.show()
Stacked Bar Chart of Best Model
# Build stacked bar chart
# Create bar_chart of confusion matrix results
bar_chart = model_search_all.loc[model_search_all.cols==('svc_four',)][["true_negatives", "false_positives",
"false_negatives", "true_positives"]]
# Stacked bar chart of correctly predicted and incorrectly predicted
stacked_bar_chart = pd.DataFrame(columns=['Correctly Predicted', 'Incorrectly Predicted'])
# Obtain totals of correctly predicted and incorrectly predicted patients
stacked_bar_chart["Correctly Predicted"] = bar_chart.filter(regex='true').sum(axis=1)
stacked_bar_chart["Incorrectly Predicted"] = bar_chart.filter(regex='false').sum(axis=1)
# Unpivot DataFrame from wide to long format
stacked_bar_chart = pd.melt(stacked_bar_chart).sort_values(by='value', ascending=False)
# # Obtain minimums of correctly predicted and incorrectly predicted patients
stacked_bar_chart["value_two"] = [bar_chart.filter(regex='true').min(axis=1).values[0],
bar_chart.filter(regex='false').min(axis=1).values[0]]
# Rename columns of long format DataFrame accordingly
stacked_bar_chart = stacked_bar_chart.rename(columns={'variable': 'Patient Outcomes', 'value': 'total',
'value_two': 'minimum'})
# Define hue and label
stacked_bar_chart['hue_label'] = list(np.where(stacked_bar_chart.total >
model_search_all.loc[model_search_all.cols==('svc_four',)][["false_positives",
"false_negatives"]].values.sum(), 'Correctly Predicted', 'Incorrectly Predicted'))
# Set colors
colors = {"Correctly Predicted": "#e66101", "Incorrectly Predicted": "#b2abd2"}
# Set figsize to size of second monitor
plt.rcParams['figure.figsize'] = [19.2,9.99]
fig, axes = plt.subplots(nrows=1, ncols=1)
fig.subplots_adjust(left=0.01, right=0.99, top=0.95, bottom=0.12)
sns_stacked_bar_plot = sns.barplot(x=stacked_bar_chart['Patient Outcomes'], y=stacked_bar_chart.total,
hue=stacked_bar_chart['hue_label'], palette=colors, dodge=False)
sns_stacked_bar_plot = sns.barplot(x=stacked_bar_chart['Patient Outcomes'], y=stacked_bar_chart.minimum,
hue=None, palette=colors, dodge=False)
for p in sns_stacked_bar_plot.patches:
print(p._height)
if p._height in stacked_bar_chart.total.values:
sns_stacked_bar_plot.annotate(int(p._height -
stacked_bar_chart.loc[stacked_bar_chart.total==p._height, 'minimum'].values[0]),
(p.get_x() + p.get_width() / 2., (p._height -
stacked_bar_chart.loc[stacked_bar_chart.total==p._height, 'minimum'].values[0])/2 +
stacked_bar_chart.loc[stacked_bar_chart.total==p._height, 'minimum'].values[0]),
ha='center', va='center', weight='bold', fontsize=30)
# Add totals above bars
sns_stacked_bar_plot.annotate(int(p._height), (p.get_x() + p.get_width() / 2., p.get_height()), ha='center',
va='center', xytext=(0, 12), textcoords='offset points', weight='bold', fontsize=30)
elif p._height in stacked_bar_chart.minimum.values:
print(p._height, p._height/2)
sns_stacked_bar_plot.annotate(int(p._height), (p.get_x() + p.get_width() / 2., p._height/2),
ha='center', va='center', weight='bold', fontsize=30)
# Set edge color to black for bars
for patch in sns_stacked_bar_plot.patches:
patch.set_edgecolor('black')
patch.set_linewidth(5)
plt.xticks(weight="bold", size=30)
plt.yticks([])
plt.xlabel('Patient Outcomes', weight="bold", size=28, labelpad=20)
plt.ylabel('')
plt.title('Prediction Results for Support Vector Machine Classifier Model #4', fontdict={"weight": "bold", "size": 28},
pad=10.0)
plt.legend(title="", prop={'weight':'bold', 'size': 20})
# Add 10 onto y to account for pad
plt.text(x=0.7, y=(stacked_bar_chart.total.max() - stacked_bar_chart.total.min())/2 + stacked_bar_chart.total.min()+10,
s = "Overall Accuracy: " + "{:.1%}".format(model_search_all.loc[model_search_all.cols==('svc_four',)]
['total_correct'].values[0]/(model_search_all.loc[model_search_all.cols==('svc_four',)]['total_correct'].values[0]
+model_search_all.loc[model_search_all.cols==('svc_four',)]['total_wrong'].values[0])),
fontdict={"weight": "bold", "size": 40}, bbox=dict(facecolor='none', edgecolor='black', pad=10.0, linewidth=3))
plt.show()
Bar Chart of Best Model
# Bar chart of confusion matrix results
bar_chart = model_search_all.loc[model_search_all.cols==('svc_four',)][["true_negatives", "false_positives",
"false_negatives", "true_positives"]]
# Rename column names accordingly
# for col in bar_chart.columns:
# bar_chart = bar_chart.rename(columns={col: col.replace("_", " ").title()})
bar_chart = bar_chart.rename(columns={'true_negatives': 'Correctly Predicted To\nNot Have Heart Disease',
'false_positives': 'Incorrectly Predicted To\nHave Heart Disease',
'false_negatives': 'Incorrectly Predicted To\nNot Have Heart Disease',
'true_positives': 'Correctly Predicted To\nHave Heart Disease'})
# Unpivot DataFrame from wide to long format
bar_chart = pd.melt(bar_chart).sort_values(by='value', ascending=False)
# Rename columns of long format DataFrame accordingly
bar_chart = bar_chart.rename(columns={'variable': 'Patient Outcomes'})
# Define hue and label
bar_chart['hue_label'] = list(np.where(bar_chart.value >
model_search_all.loc[model_search_all.cols==('svc_four',)][["false_positives",
"false_negatives"]].values.max(), 'Correctly Predicted', 'Incorrectly Predicted'))
# Set colors
colors = {"Correctly Predicted": "#e66101", "Incorrectly Predicted": "#b2abd2"}
fig, axes = plt.subplots(nrows=1, ncols=1)
fig.subplots_adjust(left=0.19, right=0.83, top=0.90, bottom=0.12, hspace=0.7, wspace = 0.25)
sns_bar_plot = sns.barplot(x=bar_chart['Patient Outcomes'], y=bar_chart.value,
hue=bar_chart['hue_label'], palette=colors, dodge=False)
# Set edge color to black for bars
for patch in sns_bar_plot.patches:
patch.set_edgecolor('black')
plt.xticks(weight="bold", size=16)
plt.yticks(weight="bold", size=16)
plt.xlabel('Patient Outcomes', weight="bold", size=18, labelpad=20)
plt.ylabel('')
plt.title('Prediction Results for Support Vector Machine Classifier Model #4', fontdict={"weight": "bold", "size": 24})
plt.legend(title="", prop={'weight':'bold', 'size': 15})
plt.text(x=1.05, y=140, s = "Overall Accuracy: " + "{:.1%}".format(model_search_all.loc[model_search_all.cols==('svc_four',)]
['total_correct'].values[0]/(model_search_all.loc[model_search_all.cols==('svc_four',)]['total_correct'].values[0]
+model_search_all.loc[model_search_all.cols==('svc_four',)]['total_wrong'].values[0])),
fontdict={"weight": "bold", "size": 22}, bbox=dict(facecolor='none', edgecolor='black', pad=10.0, linewidth=3))
plt.show()
Create Heart
plt.figure()
x = np.linspace(-2, 2, 1500)
y1 = np.lib.scimath.sqrt(1-(abs(x)-1)**2)
y2 = -3 * np.lib.scimath.sqrt(1-(abs(x)/2)**0.5)
plt.fill_between(x, y1, where=x > .7, color=colors['Incorrectly Predicted'])
plt.fill_between(x, y1, where=x <= .7, color=colors['Correctly Predicted'])
plt.fill_between(x, y2, color=colors['Correctly Predicted'])
plt.xlim([-2.7, 4.5])
correctly_predicted_patch = mpatches.Patch(color=colors['Correctly Predicted'],
label="".join([key for key, value in colors.items() if value == colors['Correctly Predicted']]))
incorrectly_predicted_patch = mpatches.Patch(color=colors['Incorrectly Predicted'],
label="".join([key for key, value in colors.items() if value == colors['Incorrectly Predicted']]))
plt.legend(title="Patient Outcomes", handles=[correctly_predicted_patch, incorrectly_predicted_patch],
prop={'weight':'bold', 'size': 16}, title_fontsize=18, bbox_to_anchor=(.97, 1.08))
# Add text to display overall accuracy if desired
# plt.text(0, -1.2, "{:.1%}".format(model_search_all.loc[model_search_all.cols==('svc_four',)]
# ['total_correct'].values[0]/(model_search_all.loc[model_search_all.cols==('svc_four',)]['total_correct'].values[0]
# +model_search_all.loc[model_search_all.cols==('svc_four',)]['total_wrong'].values[0])), fontsize=50, fontweight='bold',
# color='black', horizontalalignment='center')
plt.axis('off')
plt.show()
Go Back to Visualize Best Model
References
- Heron, M.Deaths: Leading causes for 2017 [PDF – 3 M]. National Vital Statistics Reports;68(6). Accessed November 19, 2019.
- Benjamin EJ, Muntner P, Alonso A, Bittencourt MS, Callaway CW, Carson AP, et al. Heart disease and stroke statistics—2019 update: a report from the American Heart Association. Circulation. 2019;139(10):e56–528.
- Fryar CD, Chen T-C, Li X.Prevalence of uncontrolled risk factors for cardiovascular disease: United States, 1999–2010 [PDF-494K]. NCHS data brief, no. 103. Hyattsville, MD: National Center for Health Statistics; 2012. Accessed May 9, 2019.
- Dua, D. and Graff, C. (2019). UCI Machine Learning Repository [http://archive.ics.uci.edu/ml]. Irvine, CA: University of California, School of Information and Computer Science.